Microbiome – taxonomic community and functional structure and its metabolic crosstalk with the host
The intestinal microbiome is the microbial community present in the intestinal tract. This complex microbial community is made up mainly of bacteria and covers about a many hundreds of species, although only 50 to 100 species are present in significant numbers. The community composition changes not only along the intestinal tract, but also between the mucus layer protecting the intestinal tissue and the gut lumen. This complex microbiota has a high metabolic and redundant functional capacity.
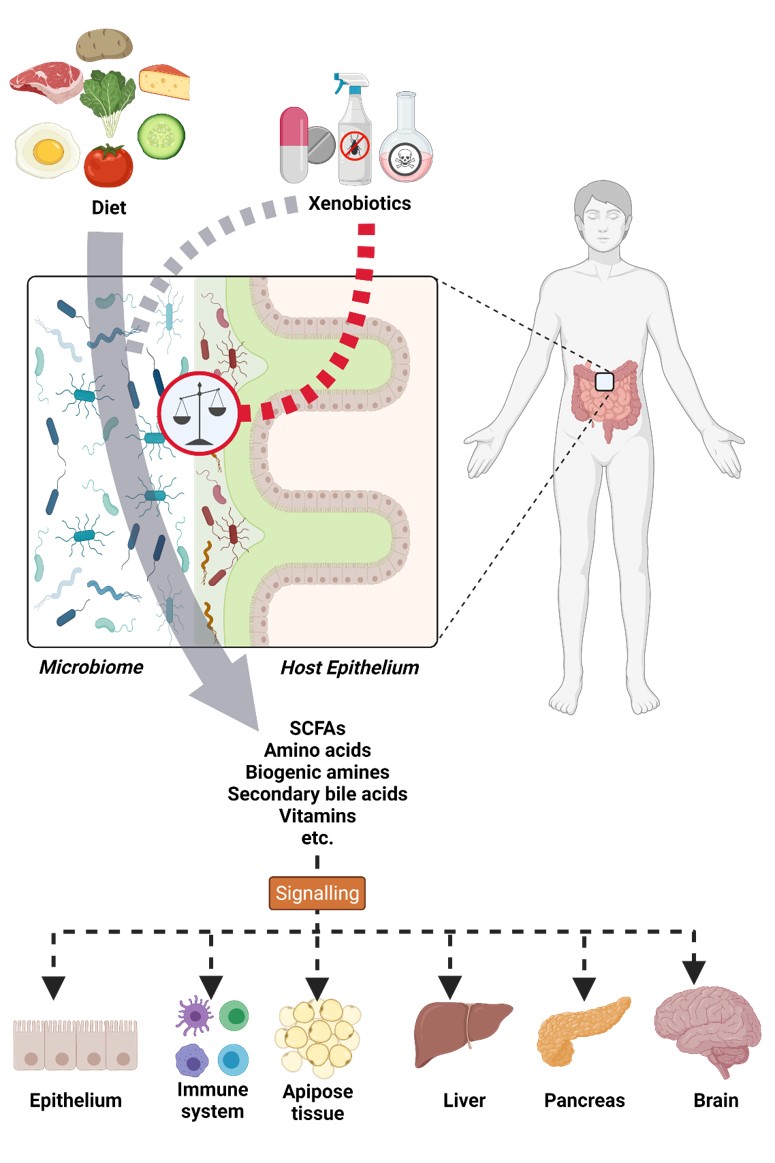
Therefore, analysing the functional interactions within the microbiome as well as between microbiome and host, and how these can change or be disrupted (dysbiosis) by external stressors such as xenobiotics (e.g. drugs, pesticides and environmental contaminants) is essential to understanding the resulting health implications. In addition, many xenobiotics can be metabolized by the microbiome and determining if this is beneficial or detrimental for the host is essential.
For this we have established a number of analytical methods including 16S rRNA gene profiling, metatranscriptomics, targeted and untargeted metabolomics as well as metabolic flux to analyse the microbiome and host samples. To integrate data we have also developed a number of software tools to automate data analysis and integration.
Though we measure samples from human participants and from animal model studies. We also have at our disposal, together with the Microbiome Biology Group, a continuous flow chemostat bioreactor system, which can be run with different intestinal microbiota models.
