16S rRNA gene profiling
16S rRNA gene profiling is a one of the most widely used methods to determine the taxonomic distribution within bacterial communities, like the intestinal microbiome. The 16S rRNA gene, which codes for a component of the bacterial ribosome, contains highly conserved and highly variable regions. The highly variable regions can be used to determine taxonomy.
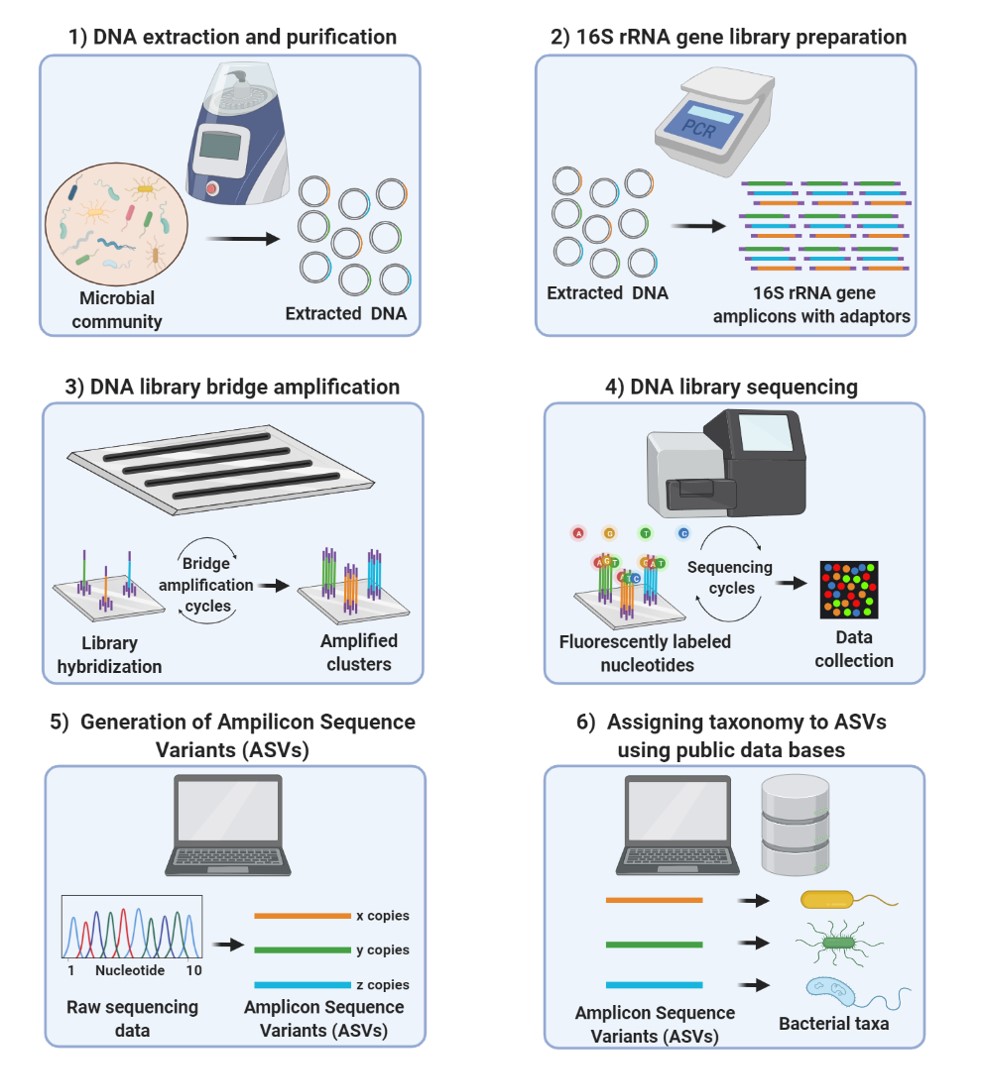
In our group, we have workflows to extract and purify DNA from fecal samples as well as to process raw sequencing data using the Dada2 algorithm on the Galaxy server at the Helmholtz Centre for Environmental Research. In addition, we have an in-house pipeline based on R-scripts for data analysis of the processed 16S rRNA gene sequencing data for biological and statistical analysis.
