Metatranscriptomics – functional insights into bacterial communities
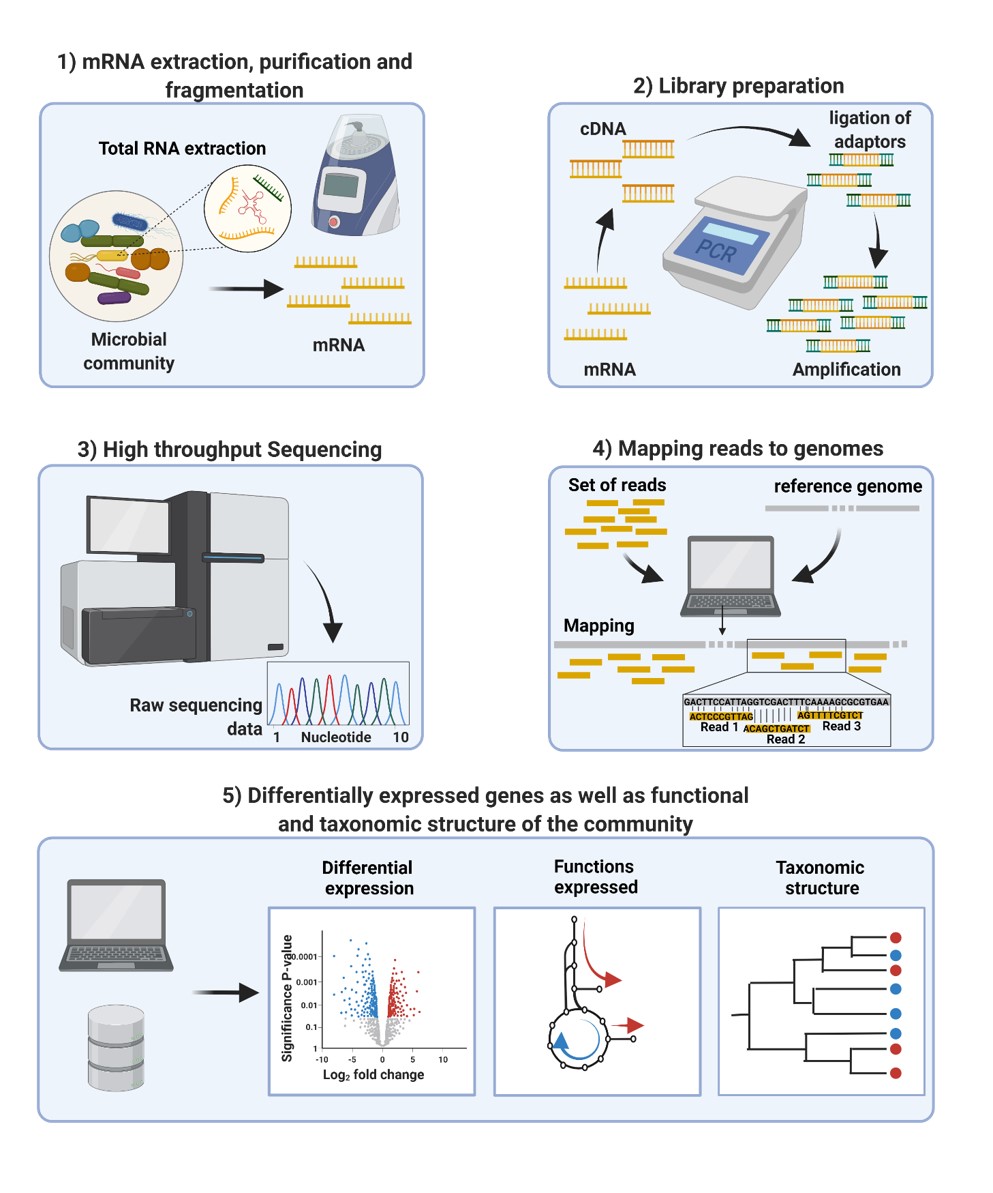
After sampling and extracting the RNA from a bacterial community, the mRNA is purified and fragmented. Using reverse transcription the mRNA fragments are converted into cDNA. This is followed by library preparation , which includes ligation of adaptors and PCR amplification, for Next Generation Sequencing (NGS). After sequencing, bioinformatic analysis matches sequencing reads to known genomes, ascribed to certain bacterial strains and the read counts per gene are a measure for the abundance of the transcript in the bacterial community. This information can be used to determine the taxonomic structure of the microbial community. In addition, transcripts can be functional annotated using databases (i.e. KEGG, EggNOG) and consequently assigned to functional pathways. Therefore metatranscriptomics is a powerful method to determine changes in taxonomy and functions expressed in the microbiome. These changes can lead to altered microbiome host crosstalk and therefore can have great health implications.
In many studies we use an continuous-flow cultivation system (bioreactor), introducing stressors and monitoring the bacteria in regards of composition and function over time. Our working group has set up a bioinformatics pipeline on the Galaxy server at the Helmholtz Centre of Environmental Research – UFZ to analyse metatranscriptomic sequencing data.
