Untargeted Metabolomics via High-Resolution Mass Spectrometry
Metabolites reflect both intrinsic biological and environmental factors. It is therefore not surprising that the aim to get a comprehensive view on the entire metabolome remains the ultimate goal. Untargeted metabolomics allows the collection of data without preexisting knowledge about the expecting metabolites, hence enabling also the opportunity to even find unknown compounds. One common method to generate untargeted metabolomics data is liquid chromatography coupled to high resolution mass spectrometry. Popular mass analyzers in that field are Orbitraps (mass-to-charge ratio is determined by the oscillation in a spindle formed trap) or time-of-flight mass analyzers (mass-to-charge ratio is determined by the time ions travel a distance).
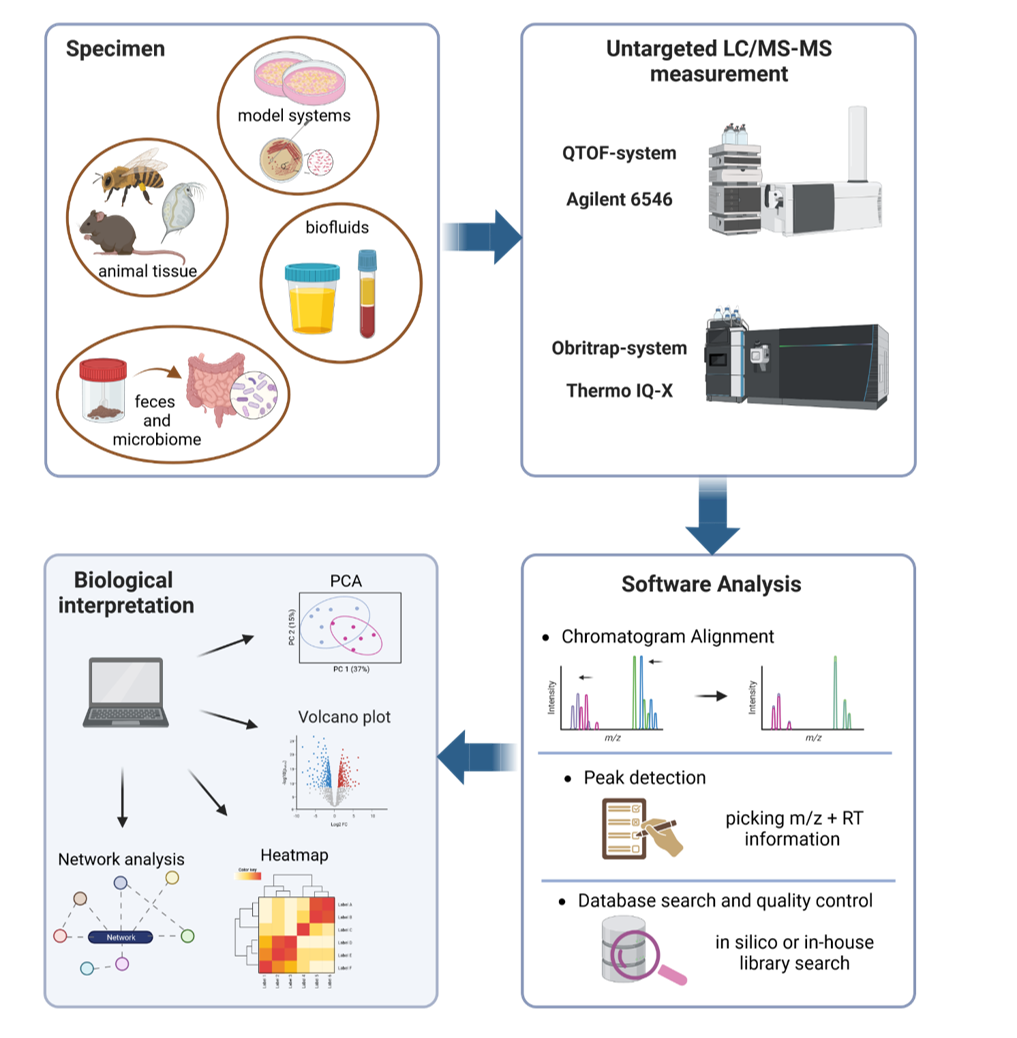
The unbiased screening to analyze the whole metabolome can be applied to a wide range of specimen including biofluids, tissue samples, environmental samples as well as microbial samples. The obtained data represent “features” (mass-to-charge ratio with a given retention time) which can be assigned to metabolites. Unfortunately, features often match with a vast number of putative metabolite structures or there may even be no candidate in the database. For accurate identification of specific features second step validation processes must follow. Since the biological interpretation hinges on the ability to identify metabolites this process remains an important challenge. To address this problem, we have started to implement a metabolite library in our working group. In doing so, we measure analytical standards from hundreds of metabolites to collect spectral information like retention time, m/z and fragment pattern. We will then be able to compare these to measured spectra from samples and increase our identification validity.
