TrophinOak and PhytOakmeter
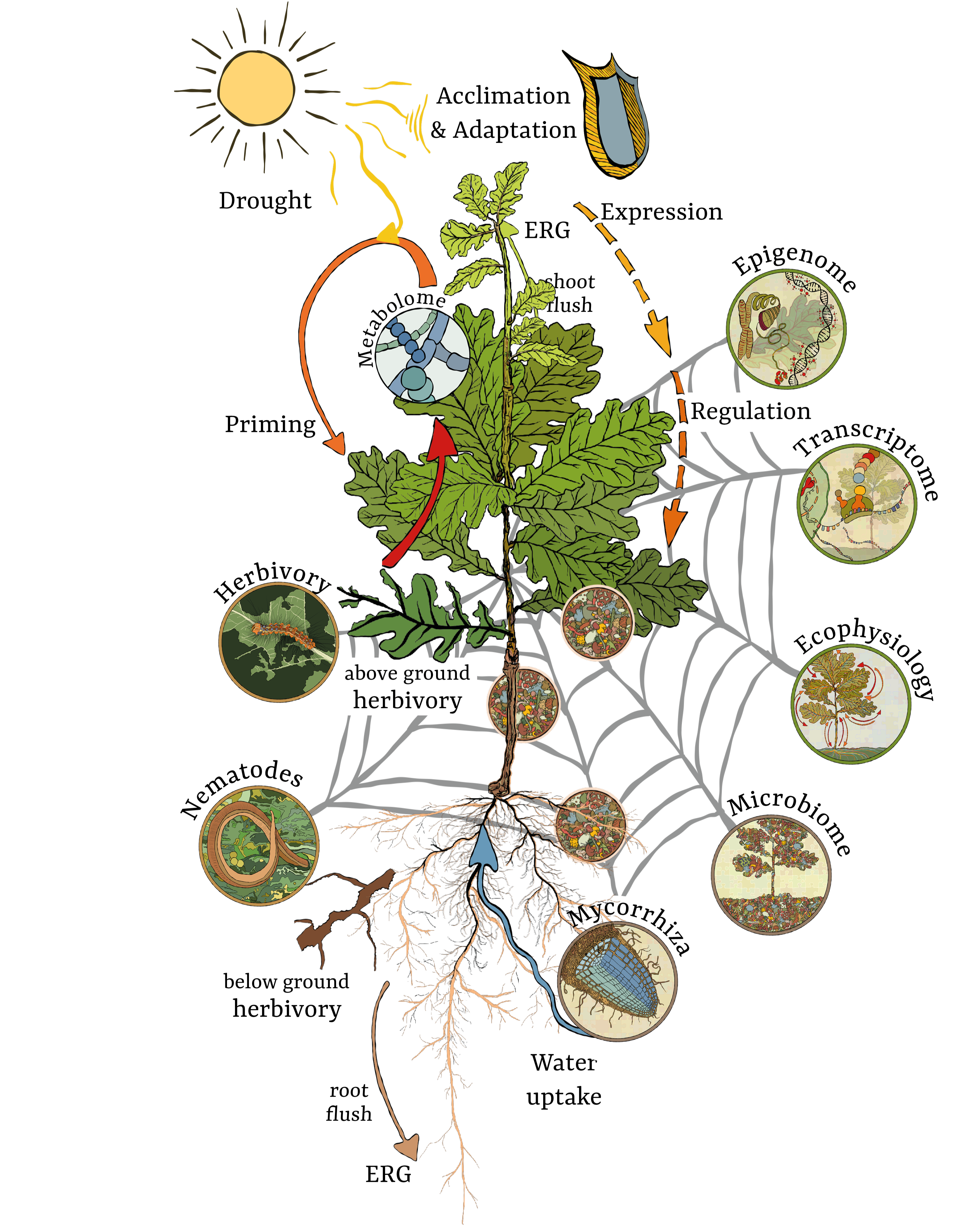
TrophinOak and PhytOakmeter focus on biotic and abiotic interactions with the clonal Oak tree DF159 under laboratory, green house and field conditions. Transcriptomic and metagenomic analyses are coupled with traits and allocation analyses in order to better understand how this important forest tree with complex multitrophic interactions can adapt under a changing environment. The TrophinOak project started in 2010 at the Helmholtz- Centre for Environmental Research - UFZ under the financial support of the DFG.

Research focus
TrophinOak
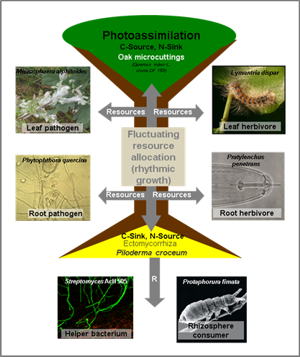
- Impact of the endogenous rhythmic growth of Quercus robur DF159 on resource allocations during multitrophic interactions under controlled conditions.
- Establishment of the OakContigDF159.1 reference library to study differential gene expression in Q. robur during biotic and abiotic interactions.
Oak Contig Database (63.7 MB)
PhytOakmeter
- Annual out planting of the Q. robur DF159 in Bad Lauchstädt and Kreinitz since 2010 to monitor the impact of the site and the tree age on soil microorganism communities in relation to the oak development and biotic above-below interactions.
- Out planting of Q. robur DF159 trees along a European geographic gradient and at a German local level under different land uses since 2013 to monitor the impact of climate change and land use management on the “soil - plant – interactor” complex.
Platform data
- Location: TrophinOak - Helmholtz Centre for Environmental Research UFZ in Halle; PhytOakmeter - Field plots in Finland, Poland, France, Bad Lauchstädt/Germany and different TERENO sites such as Kreinitz, Greifenhagen, Friedeburg, Harsleben, as well as at sites of the tree biodiversity experiment MyDiv
- Platform type: Laboratory to field experimentations with the oak clone DF159
- Research groups involved: Scientists from the Helmholtz Centre for Environmental Research, German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, Leipzig University, University of Halle, University of Tübingen, Technische Universität München, University of Göttingen, Humbold University Berlin, University of Marburg, INRA Nancy and Bordeaux in France, JGI California.
- Contact:
Prof. Dr. Mika Tarkka
,
Dr. Marie-Lara Bouffaud
- Platform projects:
DFG project - TrophinOak
(2010 – 2013)

DFG Research Unit 5571 – PhytOakmeter (2023 – 2027)

• Fernández, I., Bouffaud, M. L., Martínez-Medina, M. L., Schädler, M., Tarkka, M. T., Weinhold, A., van Dam, N. M., Herrmann, S., Buscot, F. (2024) Endogenous rhythmic growth and ectomycorrhizal fungi modulate priming of antiherbivore defences in subsequently formed new leaves of oak trees. J. Ecol. 00:1-15.
• Tarkka, M.T., Grams, T.E.E., Angay, O., Kurth, F., Maboreke, H.R., Mailänder, S., Bönn, M., Feldhahn, L., Fleischmann, F., Ruess, L., Schädler, M., Scheu, S., Schrey, S.D., Buscot, F., Herrmann, S. (2021) Ectomycorrhizal fungus supports endogenous rhythmic growth and corresponding resource allocation in oak during various below- and aboveground biotic interactions. Scientific Rep. 11, 23680
• Habiyaremye, J.d.D., Herrmann, S., Reitz, T., Buscot, F. and Goldmann, K. (2021) Balance between geographic, soil, and host tree parameters to shape soil microbiomes associated to clonal oak varies across soil zones along a European North–South transect. Environ Microbiol, 23: 2274-2292.
• Bouffaud, M.-L., Herrmann, S., Tarkka, M.T., Bönn, M., Feldhahn, L., Buscot, F. (2020) Oak displays common local but specific distant gene regulation responses to different mycorrhizal fungi. BMC Genomics 21, 399
• Habiyaremye JdD, Goldmann K, Reitz T, Herrmann S and Buscot F (2020) Tree Root Zone Microbiome: Exploring the Magnitude of Environmental Conditions and Host Tree Impact. Front. Microbiol. 11:749.
• Bacht, M., Tarkka, M.T., Fernández López, I., Bönn, M., Brandl, R., Buscot, F., Feldhahn, L., Grams, T.E.E., Herrmann, S., Schädler, M. (2019) Tree response to herbivory is affected by endogenous rhythmic growth and attenuated by cotreatment with a mycorrhizal fungus. Mol. Plant-Microbe Interact. 32 (6), 770-781
• Graf, M., Bönn, M., Feldhahn, L., Kurth, F., Grams, T.E.E., Herrmann, S., Tarkka, M., Buscot, F., Scheu, S. (2019) Collembola interact with mycorrhizal fungi in modifying oak morphology, C and N incorporation and transcriptomics. R. Soc. Open Sci. 6 (3), 181869
• Plomion, C., Aury, J.-M., Amselem, J., Leroy, T., Murat, F., Duplessis, S., Faye, S., Francillonne, N., Labadie, K., Le Provost, G., Lesur, I., Bartholomé, J., Faivre-Rampant, P., Kohler, A., Leplé, J.-C., Chantret, N., Chen, J., Diévart, A., Alaeitabar, T., Barbe, V., Belser, C., Bergès, H., Bodénès, C., Bogeat-Triboulot, M.-B., Bouffaud, M.-L., Brachi, B., Chancerel, E., Cohen, D., Couloux, A., Da Silva, C., Dossat, C., Ehrenmann, F., Gaspin, C., Grima-Pettenati, J., Guichoux, E., Hecker, A., Herrmann, S., Hugueney, P., Hummel, I., Klopp, C., Lalanne, C., Lascoux, M., Lasserre, E., Lemainque, A., Desprez-Loustau, M.-L., Luyten, I., Madoui, M.-A., Mangenot, S., Marchal, C., Maumus, F., Mercier, J., Michotey, C., Panaud, O., Picault, N., Rouhier, N., Rué, O., Rustenholz, C., Salin, F., Soler, M., Tarkka, M., Velt, A., Zanne, A.E., Martin, F., Wincker, P., Quesneville, H., Kremer, A., Salse, J. (2018) Oak genome reveals facets of long lifespan. Nat. Plants 4, 440-452
• Herrmann, S., Grams, T.E.E., Tarkka, M.T., Angay, O., Bacht, M., Bönn, M., Feldhahn, L., Graf, M., Kurth, F., Maboreke, H., Mailander, S., Recht, S., Fleischmann, F., Ruess, L., Schädler, M., Scheu, S., Schrey, S., Buscot, F. (2016) Endogenous rhythmic growth, a trait suitable for the study of interplays between multitrophic interactions and tree development. Perspect. Plant Ecol. Evol. Syst. 19, 40-48
• Maboreke, H.R., Feldhahn, L., Bönn, M., Tarkka, M.T., Buscot, F., Herrmann, S., Menzel, R., Ruess, L. (2016) Transcriptome analysis in oak uncovers a strong impact of endogenous rhythmic growth on the interaction with plant-parasitic nematodes. BMC Genomics 17, 627
• Caravaca, F., Maboreke, H., Kurth, F., Herrmann, S., Tarkka, M.T., Ruess, L. (2015) Synergists and antagonists in the rhizosphere modulate microbial communities and growth of Quercus robur L. Soil Biol. Biochem. 82, 65-73
• Herrmann, S., Recht, S., Boenn, M., Feldhahn, L., Angay, O., Fleischmann, F., Tarkka, M.T., Grams, T.E.E., Buscot, F. (2015) Endogenous rhythmic growth in oak trees is regulated by internal clocks rather than resource availability. J. Exp. Bot. 66 (22), 7113 - 7127
• Kurth, F., Feldhahn, L., Bönn, M., Herrmann, S., Buscot, F., Tarkka, M.T. (2015) Large scale transcriptome analysis reveals interplay between development of forest trees and a beneficial mycorrhiza helper bacterium. BMC Genomics 16, 658
• Lesur, I., Le Provost, G., Bento, P., Da Silva, C., Leplé, J.-C., Murat, F., Ueno, S., Bartholomé, J., Lalanne, C., Ehrenmann, F., Noirot, C., Burban, C., Léger, V., Amselem, J., Belser, C., Quesneville, H., Stierschneider, M., Fluch, S., Feldhahn, L., Tarkka, M., Herrmann, S., Buscot, F., Klopp, C., Kremer, A., Salse, J., Aury, J.-M., Plomion, C. (2015) The oak gene expression atlas: insights into Fagaceae genome evolution and the discovery of genes regulated during bud dormancy release. BMC Genomics 16, 112
• Kurth, F., Mailänder, S., Bönn, M., Feldhahn, L., Herrmann, S., Große, I., Buscot, F., Schrey, S.D., Tarkka, M.T. (2014) Streptomyces-induced resistance against oak powdery mildew involves host plant responses in defense, photosynthesis, and secondary metabolism pathways. Mol. Plant-Microbe Interact. 27 (9), 891-900
• Kurth, F., Zeitler, K., Feldhahn, L., Neu, T.R., Weber, T., Krištůfek, V., Wubet, T., Herrmann, S., Buscot, F., Tarkka, M.T. (2013) Detection and quantification of a mycorrhization helper bacterium and a mycorrhizal fungus in plant-soil microcosms at different levels of complexity. BMC Microbiol. 13, 205
• Tarkka, M.T., Herrmann, S., Wubet, T., Feldhahn, L., Recht, S., Kurth, F., Mailänder, S., Bönn, M., Neef, M., Angay, O., Bacht, M., Graf, M., Maboreke, H., Fleischmann, F., Grams, T.E.E., Ruess, L., Schädler, M., Brandl, R., Scheu, S., Schrey, S.D., Grosse, I., Buscot, F. (2013) OakContigDF159.1, a reference library for studying differential gene expression in Quercus robur during controlled biotic interactions: use for quantitative transcriptomic profiling of oak roots in ectomycorrhizal symbiosis. New Phytol. 199 (2), 529-540
• Herrmann S & Buscot F (2007) Cross talks at the morphogenetic, physiological and gene regulation levels between the mycobiont Piloderma croceum and oak microcuttings (Quercus robur) during formation of ectomycorrhizas. Phytochemistry 68:52-67.
• Frettinger, P., Derory, J., Herrmann, S. et al. Transcriptional changes in two types of pre-mycorrhizal roots and in ectomycorrhizas of oak microcuttings inoculated with Piloderma croceum . Planta 225, 331–340 (2007).
• Krüger, A., Pescaron;kan-Berghöfer, T., Frettinger, P., Herrmann, S., Buscot, F. and Oelmüller, R. (2004), Identification of premycorrhiza-related plant genes in the association between Quercus robur and Piloderma croceum. New Phytologist, 163: 149-157.
• Krüger, A., Pescaron;kan-Berghöfer, T., Frettinger, P., Herrmann, S., Buscot, F. and Oelmüller, R. (2004), Identification of premycorrhiza-related plant genes in the association between Quercus robur and Piloderma croceum. New Phytologist, 163: 149-157.