FishInspector
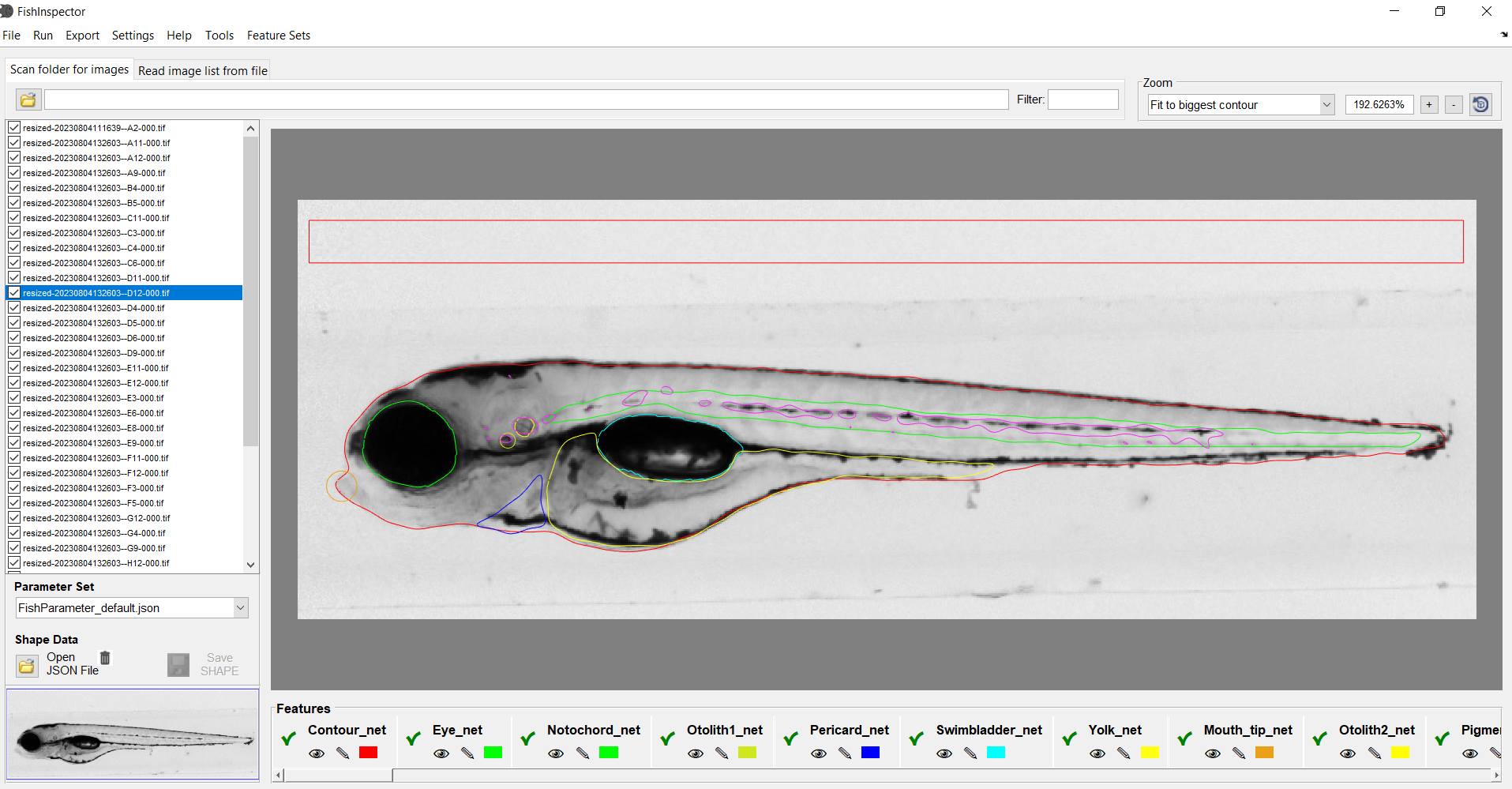
The FishInspector software allows the user-friendly and easy annotation of features in zebrafish embryo 2-dimensional images. Features are detected automatically using deep learning models but may require correction by the user (editing is implemented in the software). Both lateral and dorsal images can be analysed. The software works best with images of a defined position. When the user trains appropiate models it is possible to adopt the software to various image sources. During the installation process an appropriate version of MATLAB runtime wll be installed as well. However, no MATLAB installation is required. FishInspector has been tested on Windows 10 but may run on other windows versions as well. Mac user can use the code and or compile a version for Mac operation systems (requires Matlab license).
More details on the software and the subsequent data analysis can be found in the following publications:
Nöth, J., Busch, W., Tal, T., Lai, C., Ambekar, A., Kießling, T.R., Scholz, S. 2023. Analysis of vascular disruption in zebrafish embryos as an endpoint to predict developmental toxicity. Archives in Toxicology. Accepted manuscript.
Teixido, E., Kießling, T. R., Krupp, E., Quevedo, C., Muriana, A. & Scholz, S. 2019. Automated morphological feature assessment for zebrafish embryo developmental toxicity screens. Toxicological Sciences, 167(438–449.
Teixidó, E., Kieβling, T. R., Klüver, N. & Scholz, S. 2022. Grouping of chemicals into mode of action classes by automated effect pattern analysis using the zebrafish embryo toxicity test. Archives of Toxicology, 1-17.
Subsequent data processing
Please note that the main purpose of FishInspector is providing the coordinates of features as JSON files. For subsequent analyses appropriate workflows are required. We had been mainly interested in concentration-response analysis of fish embryos exposed to chemicals. Therefore, various KNIME workflows with embedded R-scripts were developed for data processing and analysis. The workflows are available with the corresponding publications via appropriate gitlab or github repositories (refer to the corresponding publications).
In order to run the workflows a KNIME and R installation is required. You may have to install certain additional packages in R (please open workflows in KNIME for further instructions). Our workflows have been established and tested with R version 3.4.1 but other versions may work as well.
Future Developments
We are collaborating with the Helmholtz Imaging Platform for an improved process to train semantic segmentation models for FishInspector.Acknowledgements
We acknowledge Tobias Kießling (TKS3) for implementation of our ideas into a user friendly program, establishing the MATLAB code and compiling the program as a standalone windows program. We also thank Prof. Chih Lai, St. Thomas University, MN, for support in implementing the deep learning approach.
The FishInspector software was developed with support of various research grant. Support was provided via ZFminus1 and ZF-AOP projects, both funded by the German Ministery of Education and Research (research funding scheme Alternatives to Animal Testing). Both projects were aiming at the reduction of the number of animal tests for developmental toxicity testing. Recently, further amendments of the software are supported by the European Uniion project PrecisionTox.
Contact
If you are interested in further developing the software and in collaborations, please do not hesitate to contact us.
