Partitioning of chemicals
Currently we estimate partition coefficients of neutral organic chemicals to structural proteins, plasma proteins, phospholipids and storage lipids with the pp-LFER approach (Endo et al, 2013) based on molecular descriptors from our on-line data base (LSERD). This data base contains experimental descriptors for about 8000 chemicals. For all other chemicals a QSAR provides descriptor estimates which are substantially less accurate than the experimental values. In the near future we hope to improve this situation by applying deep learning methods for descriptor estimation.
However, not only neutral organic chemicals but also ionic species have a tendency to sorb to organic matrices like proteins and phospholipids. As for neutral chemicals, this partitioning behavior of ionic species must be quantitatively understood in order to assess their toxicokinetic profile. A mechanistic understanding of the sorption behavior of organic ions is much more complex though (Henneberger & Goss, 2019). We recently performed measurements for various environmentally relevant polyfluorinated chemicals and - together with ongoing measurements for other ions - we build a larger data set that should eventually constitute the basis for further modelling.
Our final goal is a predictive model that can forecast partition coefficients with an rmse < 1 log unit. In the past we could show that the commercial software COSMOmic is able to predict the partitioning between a phospholipid bilayer and water with an rmse = 0.7 log units for monovalent acids and bases (Bittermann et al., 2016). Our goal for the future is to achieve a similar level of accuracy for the prediction of polyvalent anions and cations, zwitterions and for the partitioning to various proteins.
Deep learning applications are present in our daily lives in terms of translation programs, service hotlines, and movie suggestions on streaming portals or even in our smart homes. Deep learning is applied in image classification, face recognition, autonomous driving cars as well as in speech recognition.
Here in our department we adapt the deep learning algorithms to research questions related to a chemical’s structure to enable fast predictions of chemical properties. We compare our results to quantum-chemical based calculations which are quite reliable but also time-consuming. Deep learning algorithms should serve as fast prediction alternatives with almost comparable precision.
Contact person at UFZ: Nadin Ulrich
Biological membranes as barriers
Membranes, mainly built of phospholipid-bilayers, are the natural boundary for organisms to differentiate between exterior and interior environment–being an essential precondition for life. Due to their specific shape as a bilayer, membranes have a highly anisotropic structure including charged headgroups as well as an alkane-like hydrocarbon core. This affects the passive permeability of organic chemicals (neutral and ionic species) through membranes.
Based on an extensive literature review we have set up a model for the prediction of the passive permeability of organic chemicals through Caco-2 and MDCK monolayers. Details are found in Bittermann and Goss 2017 and the model itself can be accessed via our database (LSERD). The model is built on the widespread assumption that ionic species do not passively permeate through membranes but can only take the paracellular route. While this assumption may hold in most cases, there are cases where passive ionic permeability through membranes cannot simply be neglected. Important examples are permanent ions or uncouplers (ionophores that move protons across lipid bilayers). For their up-take or effect, ionic permeability can be the limiting process.
Permeability data on organic ions are sparse in literature. Therefore, in cooperation with Prof. Peter Pohl (Institute of Biophysics, JKU Linz, Austria), we made our own electrophysiological measurements on planar lipid bilayers. We found anionic permeabilities ranging over more than 10 log units, and were able to establish a correlation to successfully predict anionic membrane permeability based on the solubility diffusion model (Ebert et al. 2018). This correlation also holds true for perfluoroalkylic acids (manuscript submitted). These compounds exist mostly in their anionic form due to their low pKa. In cooperation with BIOVIA / Dassault Systèmes we recently extended our predictions of membrane permeability to cationic organic compounds (manuscript submitted).
Apparent permeabilities through cell monolayers such as Caco-2 are considered to be an in-vitro gold standard for assessing the uptake efficiency of drugs. Yet, in contrast to planar lipid bilayer experiments that are well explained by the solubility diffusion model, intrinsic neutral membrane permeabilities extracted from Caco-2 experiments seem not well predicted by the solubility diffusion model. Because existing experimental data between Caco-2 and planar lipid bilayers scarcely overlap, the goal of a new doctoral project will be to extend this overlap and to finally explain this discrepancy.
While passive permeability through the membrane may not always be the limiting transport process, knowledge about passive membrane permeability can help assessing whether active transport is of relevance. Also, passive membrane permeability is a key to predicting uncoupling activity.
Contact person at UFZ: Andrea Ebert and Kai-Uwe Goss
References:
Bittermann, Kai, and Kai-Uwe Goss. 2017. “Predicting Apparent Passive Permeability of Caco-2 and MDCK Cell-Monolayers: A Mechanistic Model.” PLoS ONE 12(12): 1–20.
Ebert, Andrea, Christof Hannesschlaeger, Kai-Uwe Goss, and Peter Pohl. 2018. “Passive Permeability of Planar Lipid Bilayers to Organic Anions.” Biophysj 115(10): 1931–41.
Contact person at UFZ: Andrea Ebert and Kai-Uwe Goss
In vitro - in vivo extrapolation
Bioaccumulation assessment is one criterion in the environmental risk assessment of chemicals. Usually, the bioaccumulation potential of a chemical is determined via animal testing. One promising approach that requires less animal use is the prediction of a chemical’s bioaccumulation potential using in vitro biotransformation data, because strong biotransformation prevents bioaccumulation in organisms. However, significant discrepancies are often reported when prediction results have been compared with in vivo bioaccumulation tests.
A detailed revision of the extrapolation equations showed that the commonly used extrapolation formalism leads to equations with inconsistent units indicating shortcomings in the mathematical procedure. However, even the recently presented corrected extrapolation equations (Kause, Goss 2018) could not fully resolve the discrepancies between extrapolation results and in vivo observation. In our present project, we evaluate what other reasons might explain these discrepancies and how the bioaccumulation predictions can be further improved. Currently, many processes that affect in vivo biotransformation kinetics are still not considered in the prediction models: the potentially limited delivery of the chemicals in vivo due to slow permeation of the chemical into the metabolically active cells or due to slow desorption from binding proteins in blood, the potentially decreased uptake of the chemical due to first-pass effects or the potentially higher biotransformation capacity due to the tubular architecture of blood capillaries in the liver or due to extrahepatic metabolism.
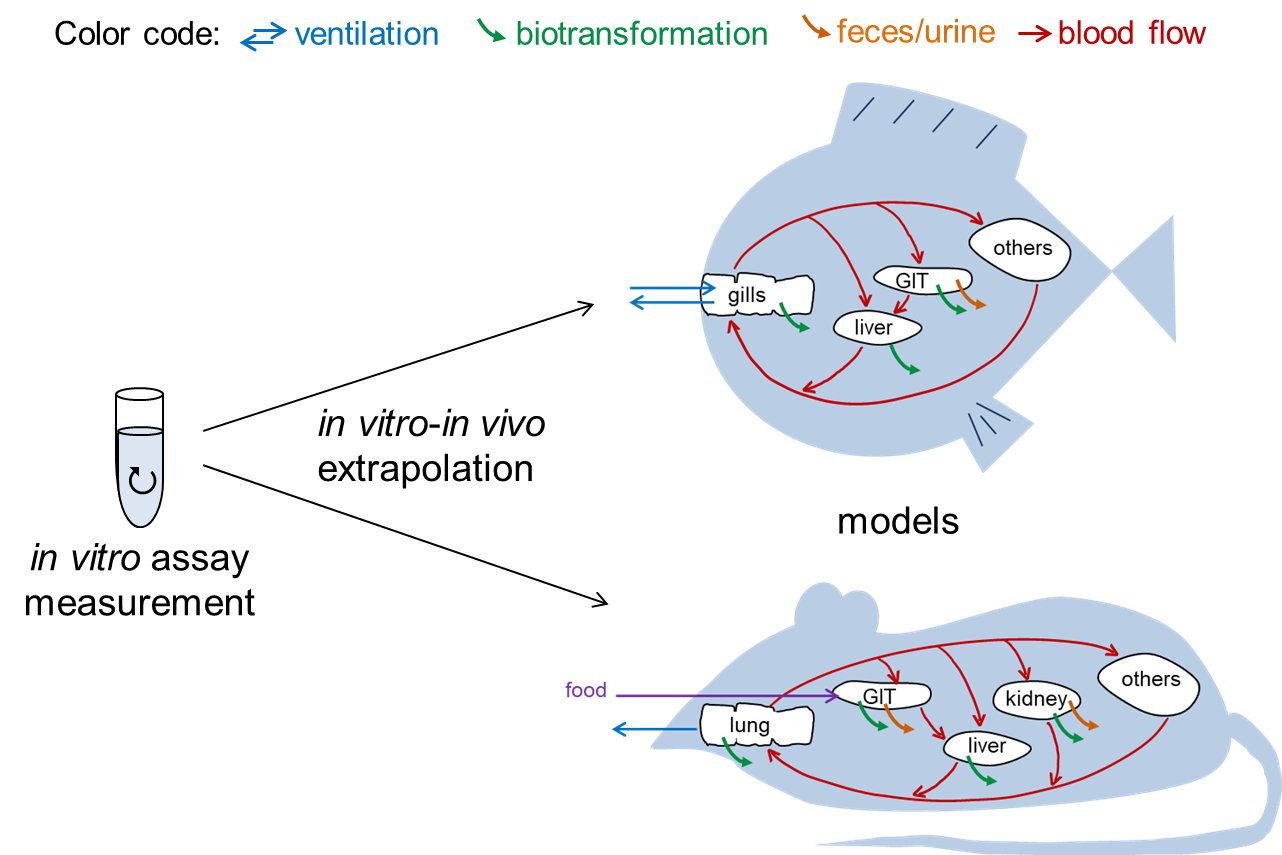
Classically, bioaccumulation evaluations mainly focused on aquatic species. However, due to the fact that aquatic species cannot represent bioaccumulation behavior in air-breathing species, methods for bioaccumulation assessment in terrestrial organisms are more and more in demand. The overall objective thus is the development of a comprehensive, easy-to-handle model for fish and rat that extrapolates in vitro biotransformation data and uses the extrapolation results for bioaccumulation prediction.
Krause, S. and K.-U. Goss, In Vitro–in Vivo Extrapolation of Hepatic Metabolism for Different Scenarios-a Toolbox. Chemical Research in Toxicology, 2018.
