Projects
Bio-Data Science
If we KNEW what we KNOW

AI for decision making
We create a data-driven platform that maps research expertise and work methodologies to optimize strategic planning and resource allocation, foster cross-functional collaboration, and enhance knowledge sharing for greater productivity and innovation.
Data Science for Chem2Bdiv

Chem-Biodiv-Networks
We integrate chemical exposure with biodiversity data and provide visual insights where chemical concentrations may impact biodiversity. We envision fostering a more sustainable relationship between human activities and the natural environment.
DeepFPlearn

AI on the Chemical Universe
We develop AI tools like deepFPlearn to predict the associations between chemicals & effects for 100s of 1000s of compounds in seconds. By including explainable AI and Quantification of Uncertainty, we enhance the credibility of the tool’s predictions for any set of compounds.
ChEOS-base

Data Integration with Graph Databases
We use a graph database to integrate chemical and toxicity information with monitoring data. It captures relationships between datasets, enabling a comprehensive analysis and providing a thorough understanding of environmental impacts on aquatic ecosystems.
ChEOS-mix
Complex Risk Drivers in Water
We use measured chemical concentrations extracted from monitoring data for chemicals measured in European surface waters from the NORMAN network data resource and integrate it with algae, daphnia, and zebrafish toxicity information to evaluate the risk of chemical mixtures on the European scale.
AI Training Data Generation

Generate transparent data for predictive toxicology
We craft advanced, adjustable pipelines for AI training data generation in predictive toxicology to ensure Reproducibility and future-proofing research. Data extraction, collection, integration, curation, and pre-processing are crucial to providing clean and precise data to ML applications.
Computational Systems Biology
Multi-omics data integration

Shaping multi-omics data for risk assessment
We are pioneering projects in multi-omics data integration, shaping their applicability for systems toxicology and risk assessment including best practices for study design and multi-omics data analysis. We are developing cutting-edge multi-omics pathway enrichment methods that embrace the complexity of biological networks and foster regulatory decision-making through pathway-based dose-response modeling.
XomeTox
Evaluating multi-omics integration for assessing rodent thyroid toxicity
The CEFIC LRI project XomeTox pioneers the use of multi-omics data in a novel, dynamic approach to unravel the complex molecular effects of toxicants, aiming to redefine our understanding of toxicity pathways. By integrating diverse omics datasets in a time- and concentration-resolved study, XomeTox seeks to discern adaptive from adverse molecular responses, paving the way for novel insights into regulatory network perturbations and toxicity prediction.
Chemical grouping
Inference of chemical grouping from processed omics data
The EFSA Tender aims to harness omics data from the CTDbase to predict chemical groupings and enhance understanding of their effects on human health and the environment. We seek to align these groupings with established CAGs, potentially facilitating the identification of additional chemicals, beyond the realm of pesticides , that might be included in these CAGs
Single-cell transcriptomics
Leveraging scRNA-Seq for toxicity assessment
We are utilizing this powerful method to uncover cell type-specific molecular responses to chemical perturbations. By analyzing data from human retina and placenta organoids as well as zebrafish embryos, we aim to shape single-cell transcriptomic analysis for regulatory applications, providing detailed insights into how chemicals impact biological systems at the cellular level.
Toxicokinetic Modeling
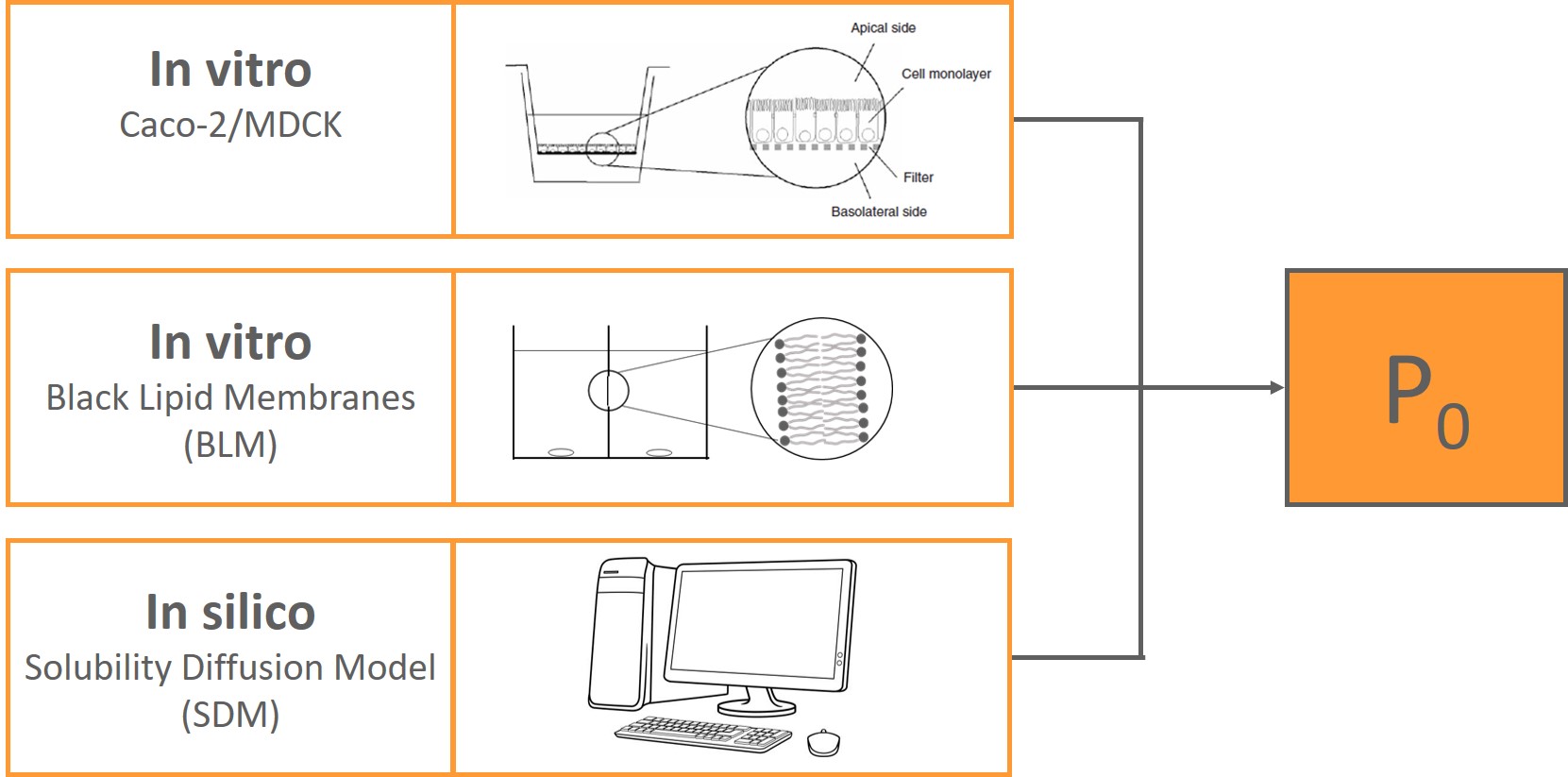
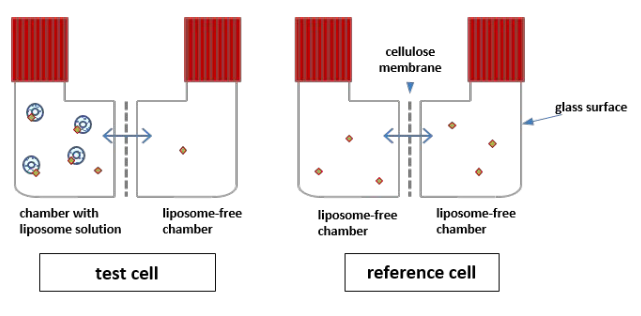
Membrane permeability predicted by the solubility-diffusion model
Using our own experiments and literature data, we show that the intrinsic membrane permeability of chemicals is well-predicted by the solubility-diffusion model, not only for artificial phospholipid membranes, but also biological Caco-2 and MDCK cell membranes.
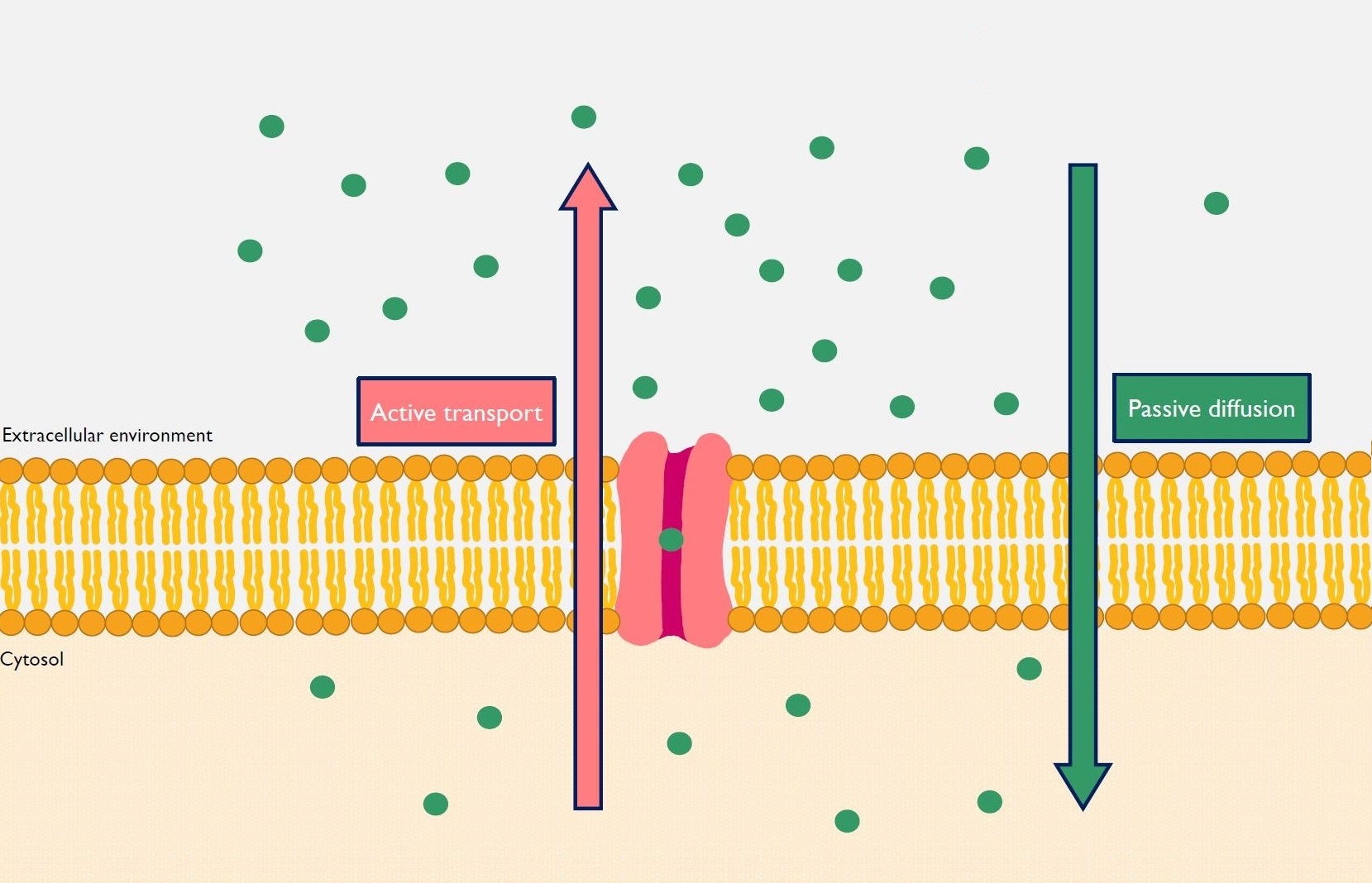
Role of active efflux in membrane permeability
We use a combined approach of mathematical modeling and transwell experiments to characterize the facilitated transport of chemicals through cellular membranes by efflux pumps.
Bioaccumulation potential of super hydrophobic chemicals
The assessment of the bioaccumulation of highly hydrophobic chemicals faces challenges, including predominant exposure through food, difficulties in conducting standard experiments, and unreliable estimation methods. Here, we aim to overcome these issues in evaluating superhydrophobic substances.
Partition coefficient prediction for neutral and ionic compounds
Our group’s expertise extends to the experimental determination and in silico prediction of partition coefficients; see the LSER database for neutral chemicals (http://www.ufz.de/lserd). For the prediction of ionic membrane/water partition coefficients, we recommend the commercial software COSMOmic.
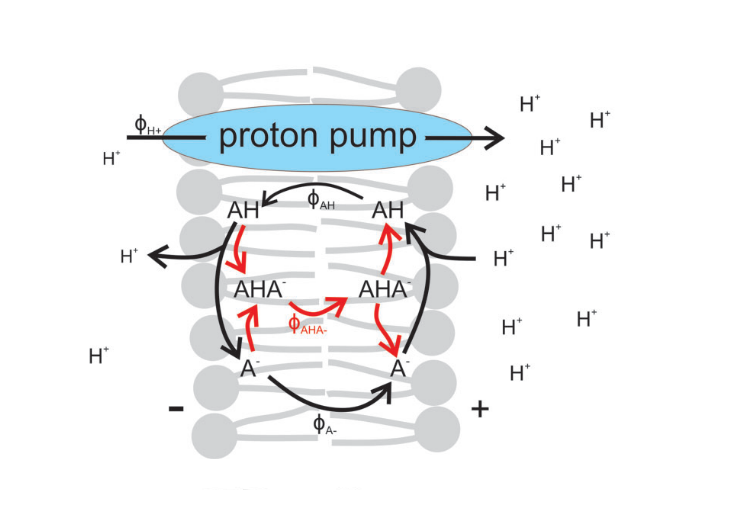
From molecular structure to uncoupling toxicity
We developed a mechanistic model to predict the toxicity of protonophoric uncoupling based on the molecule's chemical structure.
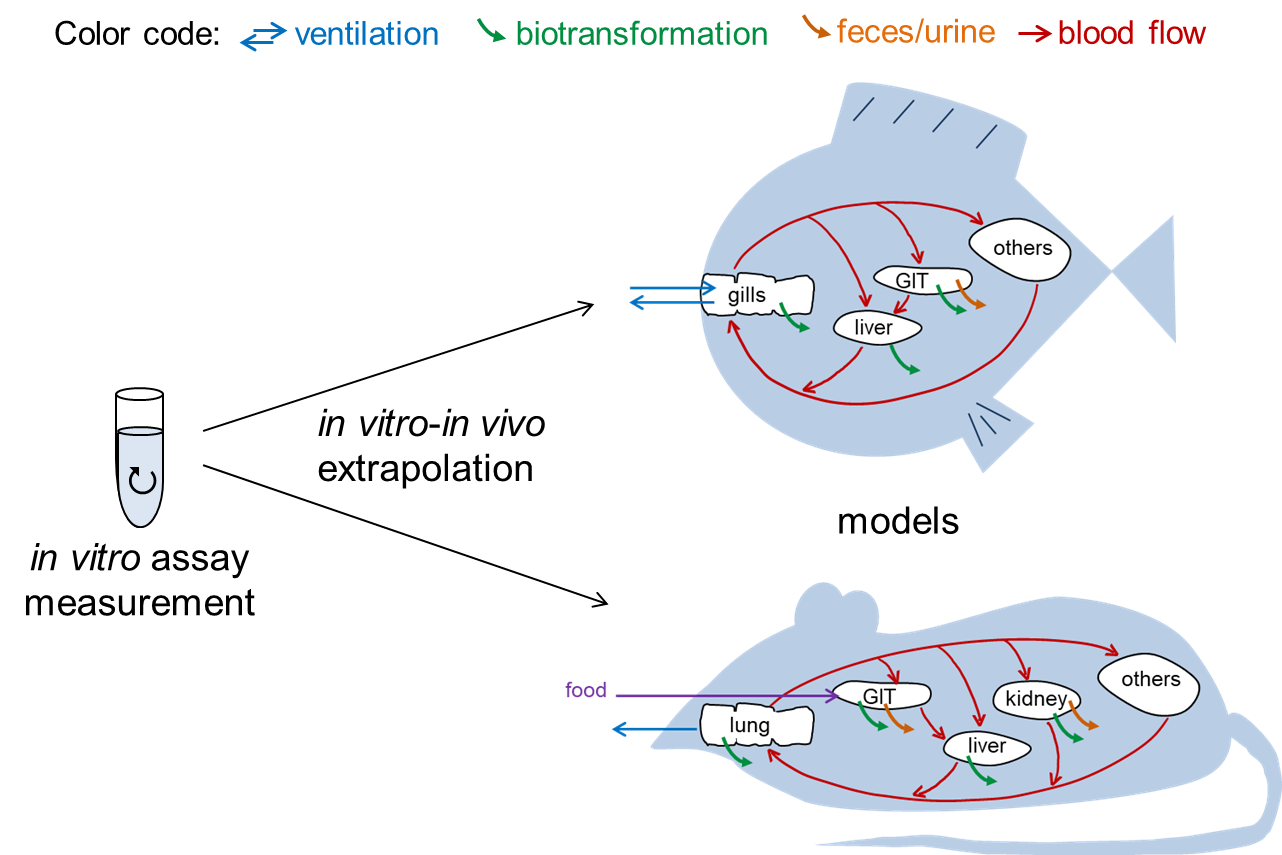
In vitro – in vivo extrapolation
We developed a comprehensive, easy-to-handle model for fish and rat that extrapolates in vitro biotransformation data and uses the extrapolation results for bioaccumulation prediction.