» Biodiversity Experiments
Funding: DFG | Project term: 2023 - 2026 (7th phase)
Web:
Biodiversity Exploratories

Microorganisms (core project 8)
#Microorganisms & Fungi
"Soil microbial diversity and community composition of grassland and forest ecosystems along land-use gradients"
The core project aims at long-term monitoring of biodiversity of soil bacteria, archaea and fungi in all 300 experimental plots as well as in the newly established field experiments (FOX, REX I+II, LUX).
Further main objectives include:- Identification and characterization of microbial key stone species employing co-occurrence network analyses, followed by selected metagenomics analyses using shotgun sequencing. This will provide better insights into functional microbial diversity.
- Identification and assessment of active bacteria by analysis of rRNA/rDNA ratios. The active bacteria are the functionally relevant proportion, since the majority of soil bacteria is inactive or dormant.
- Functional fingerprinting and reconstruction of microbial genomes from representative grassland areas (metagenomes).
The core project also provides services to contributing projects, such as long-term archiving of soil samples, standardized nucleic acid extraction and storage, next generation sequencing and support for bioinformatic analysis, provision of diversity data on soil microorganisms or organization of coordinated soil sampling campaigns.
UFZ contact:
Dr. Kezia Goldmann , Dr. Luis Daniel Prada Salcedo , Dr. Rosario Iacono , Beatrix Schnabel
Web: Microorganisms
Partners:
Leibniz Institute DSMZ - German Collection of Microorganisms and Cell Cultures (Braunschweig), Helmholtz Zentrum München, Technische Universität München (TUM)
BLD-MultiFuncDiv IV (contributing project)
#Forest & Deadwood
"BELongDead (Biodiversity Exploratories Long-term Deadwood) - Multitrophic functional diversity in deadwood"
The project investigates the decomposition of 13 temperate deciduous and coniferous tree species and the environmental drivers affecting this ecosystem process. It aims to better understand the mechanisms of species diversity and community assembly during decomposition in relation to forest management intensity and forest structure.
In frame of the BESterile (Biodiversity Exploratories deadwood Sterilization) experiment, the project additionally focuses on the mechanisms of species colonization and related processes under different forest management intensities in the early successional phase.
UFZ contact:
Dr. Julia Moll
Web: BLD-MultiFuncDiv iV
Partners:
Goethe-Universität Frankfurt am Main, Julius-Kühn-Institut (Braunschweig), Technische Universität Dresden - TU Dresden
BioNeCS (contributing project)
#Soil biology & Element cycling
"Land use and biodiversity determine the contribution of microbial necromass to soil carbon storage"
The project aims to understand how land use intensity gradients affect formation, decomposition, and stabilization of microbial necromass-derived soil organic matter by impacting the microbial community composition and its associated physiological properties.
UFZ contact:
Dr. Kezia Goldmann , Dr. Luis Daniel Prada Salcedo , Akshda Mehrotra
Web: BioNeCS
Partners:
Max-Planck-Institut für Biogeochemie (Jena), Martin-Luther-Universität Halle-Wittenberg
» Oak – an important forest tree: physiology, traits, biotic/abiotic interactions, adaption under a changing environment
Web:
PhytOakmeter

DFG Research Unit 5571 – PhytOakmeter: Using clonal oak phytometers to unravel acclimation and adaptation mechanisms of long-lived forest tree holobionts to ecological variations and climate change.
Climate change and global species loss are the major environmental threats to human well-being in the coming decades. However, fundamental knowledge is still missing about (i) the phenotypic plasticity of forest trees, (ii) about the interplay of trees with their microbiome, and (iii) about how these interactions may facilitate acclimation (regulatory changes) and adaptation based on genetic changes of trees and/or their holobiont partners. The oak holobiont represents here an assemblage of a host and the many other species living in or around it, which together form an ecological unit.
To fill this knowledge gap, the PhytOakmeter project, coordinated by Prof. Lars Opgenoorth from the University of Marburg, Germany, gathers a group of experts in population genetics, epigenetics, transcriptomics, metabolomics, biotic interactions, as well as in tree physiology and morphology. A central benefit of working with Q. robur is the availability of the DF159 clone that is readily in-vitro propagated in large numbers. This resource allows us to exclude genetic variability of the tree host in order to disentangle the role of the holobiont partners in the acclimation and adaptation processes of the oak holobiont.
The UFZ Department of Ecology of Agroecosystems is involved in the following subprojects:
Central coordination project C2 – Experimental Platforms and Plant Material
"Setting up and maintaining experimental resources"

UFZ contact: Dr. Marie-Lara Bouffaud ,
Prof. Mika Tarkka
Web: Central coordination project C2
Subproject 2 – Holobiont transcriptomics
"Holobiont acclimation and adaptation to global change as revealed by the transcriptome"
UFZ contact: Dr. Marie-Lara Bouffaud
Web: Subproject 2
Subproject 4 – Microbiome
"Cross talk between environmental conditions, oak tree performance, and the microbiomes of their soil, endo- and and phyllosphere"

UFZ contact: Dr. Kezia Goldmann ,
Dr. Luis Daniel Prada-Salcedo ,
Camilo Andres Quiroga Gonzalez
Web: Subproject 4
Subproject 5 – Ectomycorrhiza
"Adaptation and acclimation of the clonal oak's ectomycorrhizal fungi to biotic and abiotic environmental factors"
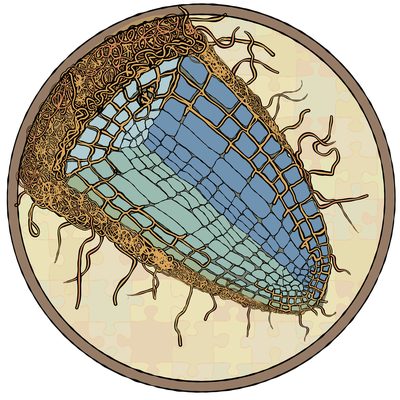
UFZ contact: Prof. Mika Tarkka ,
Erik Teutloff
Web: Subproject 5
Partners: Philipps-Universität Marburg, Universität Freiburg, Universität Leipzig, German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, Eidg. Forschungsanstalt für Wald, Schnee und Landschaft (WSL) in Birmensdorf (Schweiz) (more details see
here
)
» GCEF – Global Change Experimental Facility: exploring ecosystem processes under climate and land use-specific changes
Funding: UFZ (PhD college) | Project term: 2024 - 2027

„Coupling circular economy principles with environmental hazard management for liquid manure-based sustainable agriculture“
SmartManure develops solutions for on-farm liquid manure management as an essential path to sustainable agriculture. It will combine advanced technologies with ecosystem engineering to transform on-farm liquid manure into an environmentally friendly economic resource, while promoting sustainable agriculture by establishing more resilient ecosystems.
Within four PhD projects, SmartManure will provide a technical concept for the economic use of liquid manure during on-farm storage providing three products simultaneously: methanol and heavy metals for sale to industry, and safe fertilizer low in antibiotics for direct use on farm. It will further assess impacts of liquid manure application on crop performance, microbial agroecology and soil health.
Web: SmartManure

Platform: GCEF

The Department of Ecology of Agroecosystems (AECOL) is involved in two PhD projects:
- PhD3
“Cover crops for resilient soil microbiomes and improved nutrient retention”
The project will increase soil microbiome resilience towards manure-based hazards and simultaneously retain manure-based nutrients in the soil by growing cover crops during intermittent seasons.
Contact:
Dr. Anja Worrich (Dep. AME), Lieke Lipsch (PhD, Dep. AME), Dr. Sara König (Dep. BOSYS), Dr. Luis Daniel Prada Salcedo (Dep. AME, AECOL), Ass. Prof. Dr. Marie Muehe (Dep. AME) - PhD4
“Intercropping for mitigating manure-based hazards to corn production”
The project will minimize contaminant impacts on cash crops by intercropping in liquid manure-based agriculture, providing high-quality, abundant food and intercrop-based bioenergy.
Contact:
Lena Mellin (PhD, Dep. AECOL), Ass. Prof. Dr. Marie Muehe (Dep. AME), Dr. Martin Herzberg (Dep. ANA)

Funding: UFZ | Project term: 2022 - 2025
GLIMPSE Research consortium: „Global change impacts on microbiota-plant-soil processes relevant for water and matter cycling in agricultural ecosystems“
- subproject P2
"Interacting effects of global change drivers on the fauna and microbiome of agricultural soils and the consequences for related ecosystem processes"
The project (P2) will assess the interacting effects on functional traits of soil microbiota and related processes ensuring ecosystem functions and resilience. This specific project aims at experimental investigations of combined land use and global change effects on the structure and functions of the soil fauna as well as of the microbial community.
Contact:
Dr. Thomas Reitz (PI), Dr. Evgenia Blagodatskaya , Dr. Martin Schädler (Dep. BZF, UFZ), Lena Philipp (PhD) - subproject P3
"Root-Soil Interactions"The project (P3) focuses on the link between root systems and soil structure with implications for water and nutrient fluxes.
Contact:
Dr. Steffen Schlüter (PI, Dep. BOSYS), Prof. Dr. Mika Tarkka , Prof. Dr. Doris Vetterlein (Dep. BOSYS)
» Further Projects
Funding: Helmholtz Association, Innovation Pool of the Research Field Earth and Environment | Project term: 2025 - 2027
Landscapes as Carbon Sponges (CSponge) – Towards long-term net-negative land use
UFZ-Contact:
Dr. Evgenia Blagodatskaya
(Dep. AECOL), Dr. Oliver Lechtenfeld (Dep. EAC), Dr. Christian Siebert (Dep. CATHYD)
Partners:
Helmholtz Centre Potsdam, GFZ German Research Centre for Geosciences; Helmholtz Centre for Polar and Marine Research, Alfred Wegener Institute (AWI)
Funding: BMBF - Scientific and Technological Cooperation (STC) with Ukraine
Project term: 2024 - 2026
Microbial biologically active metabolites as a biotechnology tool to improve crop productivity and soil health (MicroMet)
UFZ-Contact:
Dr. Evgenia Blagodatskaya
(Dep. AECOL),
Prof. Dr. Mika Tarkka
(Dep. AECOL), Dr. Hryhoriy Stryhanyuk (Dep. TECH), Dr. Nadiiia Yamborok (Dep. TECH)
Partners:
D.K. Zabolotny Institute of Microbiology and Virology of the National Academy of Sciences of Ukraine
Funding:


Project term: 2023 - 2028 | Web: MicroPLUS
Micro- and Meso-PLastics: Identification and Utilization in Soil (MicroPLUS)
Considering a very broad range of bio-based and bio-degradable plastics used in the modern agriculture and numerous plastic degradation pathways by large number of microorganisms there is an urgent need to find efficient ways for plastics identification in the environment, investigate the mechanisms and pathways of plastics transport and decomposition in soil and water and its consequences for carbon stocks and soil health.
This project contributes to the Microplastics Competence Cluster (MPCC) at the UFZ that bundles the expertise of diverse research groups at UFZ and beyond about different aspects of plastics and associated chemicals in the environment.
MPCC
Contact:
Dr. Evgenia Blagodatskaya
(Dep. AECOL), Dr. Matthias Schmidt (Dep. TECH), Dr. Hryhoriy Stryhanyuk (Dep. TECH)
Funding: China Scholarship Council (CSC) | Project term: 2023 - 2027
Web: CSC

Project
"Effects of impaired soil P and N stoichiometries on plant symbioses with microorganisms"
Phosphorus (P) often limits the primary productivity of natural and agroecosystems because of its low availability and mobility in most soils. In response to low P environments, vascular plants have evolved a range of phosphorus acquisition strategies, such as forming root hairs or increasing the root surface by branching. Support to plant P acquisition is obtained from phosphate solubilizing microorganisms that solubilize mineral and organic P, and arbuscular mycorrhizal fungal (AMF) mycelium that provide the mycorrhiza pathway for phosphate uptake that reaches outside the P limitation zone around the roots. The microorganisms may thus account for up to 90% of the total P requirement of the plant. Microorganisms also have a central role in soil nitrogen (N) cycling and plant N uptake. For instance, AMF can acquire N from both mineral and organic N sources and transfer some of this N to their host plants, suggesting a central role in N acquisition in N poor soils. The specific strategies for plant phosphorus acquisition under nutrient addition-induced soil stoichiometric N:P imbalance remain unclear. Based on this, we investigate the following:
Under conditions of different soil N:P stoichiometries, induced by controlled nitrogen and phosphorus fertilizations:- How do the AMF and root architecture respond?
- How do the responses of AMF and phosphate solubilizing microorganisms interact?
- How are root pathways, mycorrhizal pathways, and leaf reabsorption pathways balanced?
- What are the mechanisms underlying the response and interaction between AMF and mycelial bacteria?
- How is this related to the bacteria involved in nitrogen cycling?
Contact:
Prof. Dr. Mika Tarkka , Dr. Kezia Goldmann , Dr. Thomas Reitz , Cheng Peng
Platform:
Static Fertilization Experiment Bad Lauchstädt
Funding: project executing agency of the BMEL: Fachagentur Nachwachsende Rohstoffe e.V. (FNR), Waldklimafonds | Project term: 2023 – 2025 |
Web:
BBZE-Wald

Subproject 2
“Analysis of the biodiversity and ecological functions of soil fungi and soil bacteria”
Networks of soil organisms and plants are key drivers of biogeochemical cycles in terrestrial ecosystems. Soil communities of fungi, bacteria, archaea, and other protozoa are important for functions such as C storage, tree resilience to climate change, and matter turnover in forest soils. There is a nationwide monitoring of tree vitality and soil physicochemical condition with the Soil Condition Survey in Forests (BZE). However, soil biology has not yet been considered. An extended systematic monitoring can help to understand relationships between site conditions, soil organisms and their functions. This project therefore aims to link the comprehensive data of the BZE with newly collected data on structural and functional biodiversity of soil microorganisms.
UFZ contact:
Dr. Evgenia Blagodatskaya
Partner:
Johann Heinrich von Thünen-Institut (Braunschweig) – Institut für Waldökosysteme
DFG Priority Program 2089 “Rhizosphere Spatiotemporal Organization – a Key to Rhizosphere Functions”
Funding: DFG (TA 290/5-1) | Project term: 2018 - 2021 (first phase)
2022 - 2025 (second phase)
Web:
SPP - Rhizosphere
 The Rhizosphere Priority Program aims at the identification of patterns in space and time in the volume of the soil affected by living roots, the rhizosphere. Its major goal is the explanation of the mechanisms behind and feedback processes between the soil, soil microorganisms and the plants during rhizosphere establishment. The overall hypothesis of the Priority Program is that resilience emerges from self-organized spatiotemporal pattern formation in the rhizosphere.
The Rhizosphere Priority Program aims at the identification of patterns in space and time in the volume of the soil affected by living roots, the rhizosphere. Its major goal is the explanation of the mechanisms behind and feedback processes between the soil, soil microorganisms and the plants during rhizosphere establishment. The overall hypothesis of the Priority Program is that resilience emerges from self-organized spatiotemporal pattern formation in the rhizosphere.
contributing project P8 - Root Genes I and II
“Spatial and temporal analysis of maize root gene expression patterns as a tool to elucidate how the maize interacts with rhizosphere microbiome and the environment”
The central aim of this associated project in frame of the SPP 2089 is to reveal mechanisms how maize roots adapt to changes in soil environment, and how the adaptations contribute to rhizosphere processes at a single root type and at root system levels. Based on the special importance of substrate, observed in the first phase, the project aims at a more precise understanding of the substrate induced changes in gene expression. It concerns soil compaction that is an important factor for agricultural production and. Carbon allocation that represents an important modifier of plant defense response and rhizosphere functioning, and is also related to maize gene expression. Our data suggest that the changes related to substrate and soil depth may be related to differences in soil moisture. Drought events are predicted to increase in frequency and intensity and cause a challenge for agricultural production, and this aspect is included to the research program. To address the impacts of substrate, soil depth and maize genotype at a higher scale, the annual development of the maize field is included in the frame of Complex Sampling among the participants of the priority program.
Contact:
Dr. Mika Tarkka , Minh Ganther (first project phase), Henrike Würsig (second project phase)
Partners:
members of associated subprojects within the DFG Priority Program SPP 2089
contributing project P13 - Microzym I and II (Kinetics & Microzymography)
“Microbial life in the rhizosphere: kinetics and visualization of the functions”
The general objective of Microzym is visualization and quantitative analysis of biochemical processes in the rhizosphere enabling to link microbial functional traits with localized process rates in space and time.
Specific objectives:- Development and modification of methods for spatial analysis of distribution of root and of microbial activity parameters considering soil structure at the aggregate level. For this objective, the range of methods developed during first stage of PP (e.g., time-lapse zymography, oxidative zymography, micro-zymography, aminography) will be further extended (to e.g., oxidative micro-zymography, micro-aminography and amino-mapping) and localized micro-sampling at the level of individual aggregates will be applied for spatial and temporal identification of soil and root properties responsible for microbial activities and functions.
- Comparison of microbial feedback to unfavorable environmental conditions in the rhizosphere of two maize genotypes. For that, we aim to relate the localization of enzymatic activity with microbial growth parameters at the epi-centrum, at the boundary and outside the rhizosphere hotspots under limitation by nutrients and water.
- To reveal spatial specificity of organic substrates transformation and turnover at the rhizosphere – detritusphere interfaces as regulated by the rates of root detritus decomposition.
- To study a shaping effect of soil profile formation induced by maize monoculture on microbial and enzymatic functional traits in the rhizosphere.
Contact:
Dr. Evgenia Blagodatskaya , Negar Ghaderi Golezani (first project phase), Zeeshan Ibrahim (first project phase), Maria Martin Roldan (second project phase)
Partners:
members of associated subprojects within the DFG Priority Program SPP 2089
Funding: DFG | Project term: 2022 - 2025 | Web:
SPP - SoilSystems
 The DFG joint research program SoilSystems aims to integrate a thermodynamic description of the soil system in order to gain a systemic view on energy and matter fluxes and their interactions with living and non-living soil components. This will enable to elucidate dynamic biogeochemical processes, boundary constraints and performance limits, and to identify optimally approaches allowing to describe the complex energy-driven soil systems in much simpler terms.
The DFG joint research program SoilSystems aims to integrate a thermodynamic description of the soil system in order to gain a systemic view on energy and matter fluxes and their interactions with living and non-living soil components. This will enable to elucidate dynamic biogeochemical processes, boundary constraints and performance limits, and to identify optimally approaches allowing to describe the complex energy-driven soil systems in much simpler terms.
contributing project P5 - MICROHEAT
“Heat dissipation and matter turnover in the course of microbial succession constrained by substrate and oxygen availability”
Current challenge to increase sequestration of soil carbon (C) - the largest and most important terrestrial C stock - requires understanding the processes of carbon losses during microbial decomposition of diverse organic compounds, which serve as a substrate to obtain C, energy and nutrients for microbial metabolism.
The project Microheat considers that transformation of organic substrates in soil occurs in the course of multi-stage microbial succession through a sequence of decomposition stages. The methodological novelty and challenge of Microheat, therefore, is in estimation of the anaerobic soil volume fraction via application of X-ray CT to study the effect of oxygen diffusion constraints on C and energy use efficiency (CUE and EUE). The peculiarity of Microheat is in its conceptual view linking matter (CO2) and energy (heat) losses to microbial growth and to the range of enzyme-mediated reactions related to various stages of substrate decomposition. The Microheat project will make a promising contribution to elucidating regulatory mechanisms of energy and matter turnover based on the microbial trade-offs between rate and efficiency of organic compounds decomposition in soil.
Contact:
Dr. Evgenia Blagodatskaya , Dr. Thomas Reitz , Dr. Steffen Schlüter (Dep. BOSYS, UFZ), Fatemeh Dehghani Mohammad Abadi
Partners:
members of associated subprojects within the DFG Priority Program SPP 2322
Funding: DFG | Project term: 2022 - 2025 | Web:
SPP - DECRyPT
The central scientific objectives of the Priority Programme DECRyPT are to obtain a deep and more predictive understanding of plant-microbiota associations and to develop pioneering reductionist approaches towards a molecular understanding of plant microbiota functions. In this Priority Programme we elucidate genetic factors underlying plant microbiota establishment, test presumed community adaptation in ecological contexts and define community-associated emergent properties. Computational and genomic tools will guide hypothesis testing and the design of microbiota reconstitution experiments in controlled environments.
contributing project: DTsyncom
“A drought tolerant synthetic bacterial community for barley”
Drought events will increase in frequency and intensity and thus increasingly become a challenge for agricultural production. It is well known that plants respond to drought with changes in the plant-associated microbiome, and the adapted microbiome can thus contribute to increasing plant fitness. The proposed project investigates interaction mechanisms between synthetic bacterial communities and barley under drought stress. Bacterial and plant gene expressions, as well as measurements of barley drought tolerance, will be the focus.
Contact:
Dr. Mika Tarkka , Dr. Anna Heintz-Buschart (Universität Amsterdam- UVA), Dr. Thomas Reitz , Linda Rigerte
Partners:
members of associated subprojects within the DFG Priority Program SPP 2125
Funding: China Scholarship Council (CSC) | Project term: 2021 - 2024
Web: CSC

Project
"Coupling zymography with enzyme kinetics to visualize microbial community dynamics in the rhizosphere"
Contact: Dr. Evgenia Blagodatskaya , Guoting Shen