Omics-based grouping of chemicals -
Inference of chemical grouping from processed OMICS data in CTD
The EFSA project embarks on an innovative approach to chemical risk assessment, utilizing the rich omics data from the Comparative Toxicogenomics Database (CTD) to systematically group chemicals according to their molecular effects. This initiative aims to refine the CGPD tetramer calculation method, originally proposed by CTD, to identify chemicals sharing common molecular responses. Structured around a series of meticulously planned tasks, the project is designed to harness the extensive range of chemical data within CTD, including pesticides, pharmaceuticals, and plasticizers, for a more comprehensive chemical grouping.
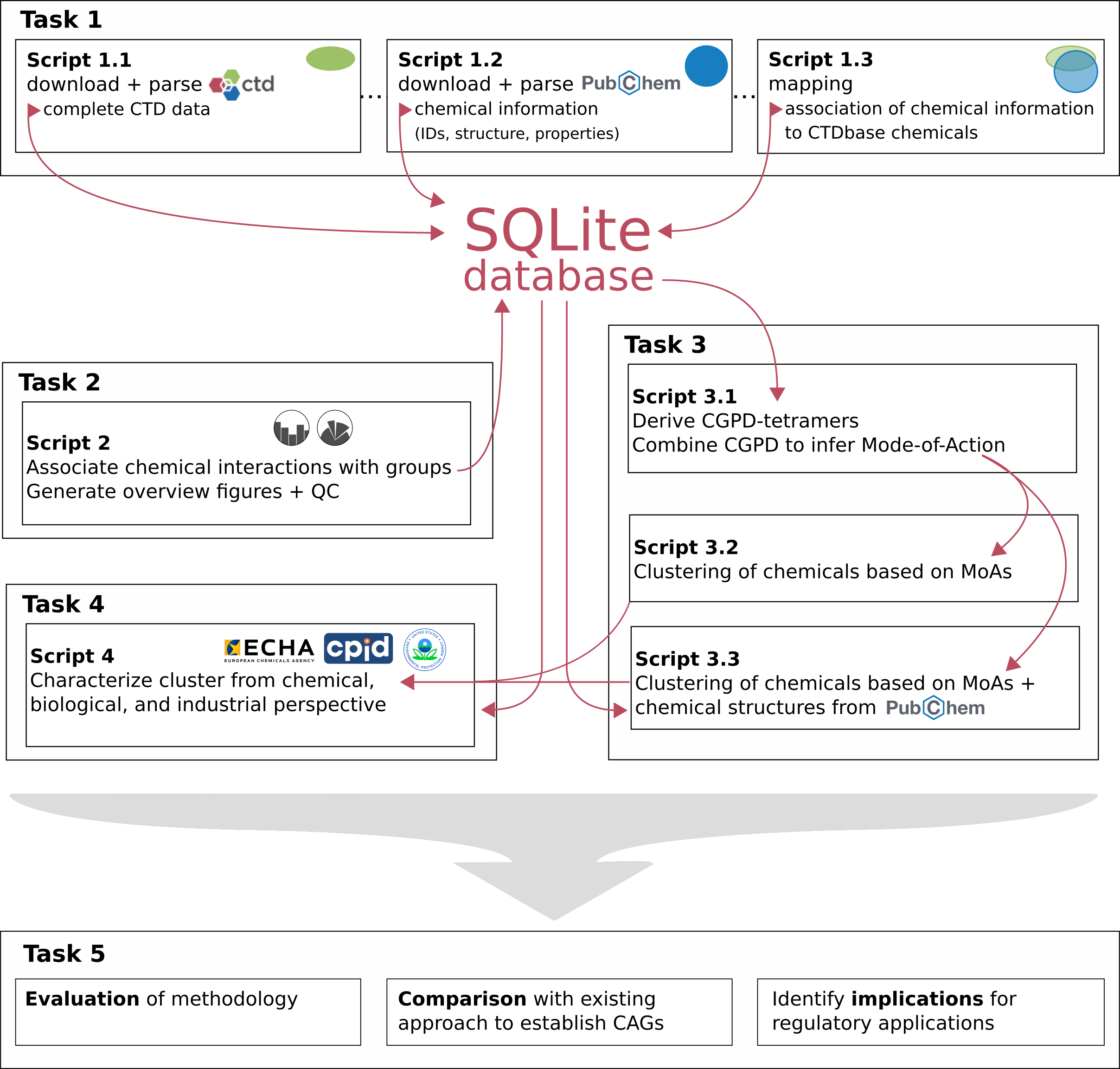
In its second phase, the project focuses on comparing these newly established chemical groups with the existing Cumulative Assessment Groups (CAGs) for pesticides, employing a holistic approach that encompasses a broader spectrum of chemicals. This comparative analysis seeks to validate the grouping methodology while also aiming to expand the scope of CAGs to include a wider variety of chemical types. By integrating this broader range of chemicals, the project aspires to enhance the understanding of chemical safety, providing a more robust framework for assessing the potential health and environmental impacts of various compounds.
Reproducibility of work
Reproducibility and FAIR public access to data and code is generally a major aim of our work. The working environment will be provided in a Singularity container to ensure reproducibility on any given machine. All necessary programming languages (Python and R), the database environment (SQLite database), and all required tools and libraries (as conda environment) will be included in the container file. Scripts and tools that will be developed during the project will be made publicly available on code sharing platforms like GitLab and will also be part of the Singularity container. Through a continuous integration workflow, we are able to ensure stable versioning of our container, higher test reliability, and faster release rates.
The combination of publicly available code, a publicly available and ready-to-go working environment in the form of a Singularity container combined with FAIR accessible data guarantees the highest level of reproducibility. This approach allows anyone to rerun our analyses using exactly the software environment we used without the need to install any software by just deploying the Singularity container.
Funded by: European Food Safety Authority
(EFSA)
Duration: 02/2023 - 02/2024
Related own publications:none yet
