P17 - Microbial networks
Self-organized microbial networks in soil aggregates and their dynamics in the root soil continuum
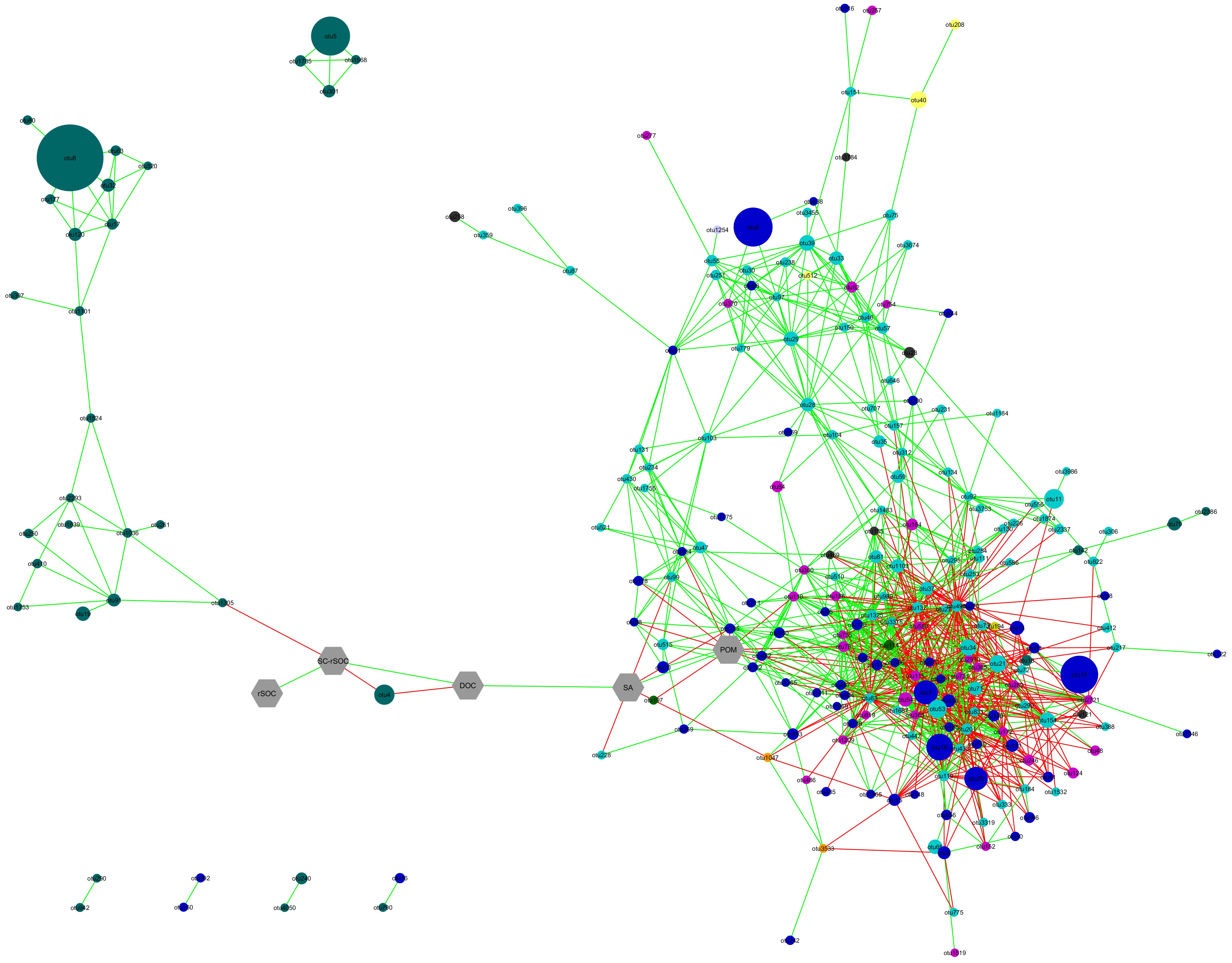
Emerging properties of microbial communities are a result of the functional potentials of their constituting members. The specific conditions in the rhizosphere select for microorganisms that, on one side, support plant growth in return for receiving energy rich carbon sources from rhizodepositions, and, on the other side, compete with plant roots for growth nutrients, especially soil nitrogen (N) and phosphorous (P). These can be considered as positive and negative feedbacks between the plant and the rhizosphere microbial community, respectively. During the 1st phase of this SPP 2089, we established a protocol for extracting from individual aggregates and characterized the bacterial community composition by DNA 16S rRNA PCR amplicon sequencing. Networks between bacterial community members obtained with aggregate DNA indicated a strong effect of root hairs not seen with alpha-diversity measurements. For the 2nd phase we take this approach further and characterize the aggregates at the level of their microbial functional potentials, based on studying the diversity of alternative genetic versions of microbial key functions contributing to soil N transformation and P mobilisation. We will combine our molecular microbial community results with information on the aggregates’ 3D architecture and other physico-chemical aggregate properties, as provided from SPP partners. In addition, functional adaptations will be studied by shotgun metagenomics of individual soil aggregates.
Outcome
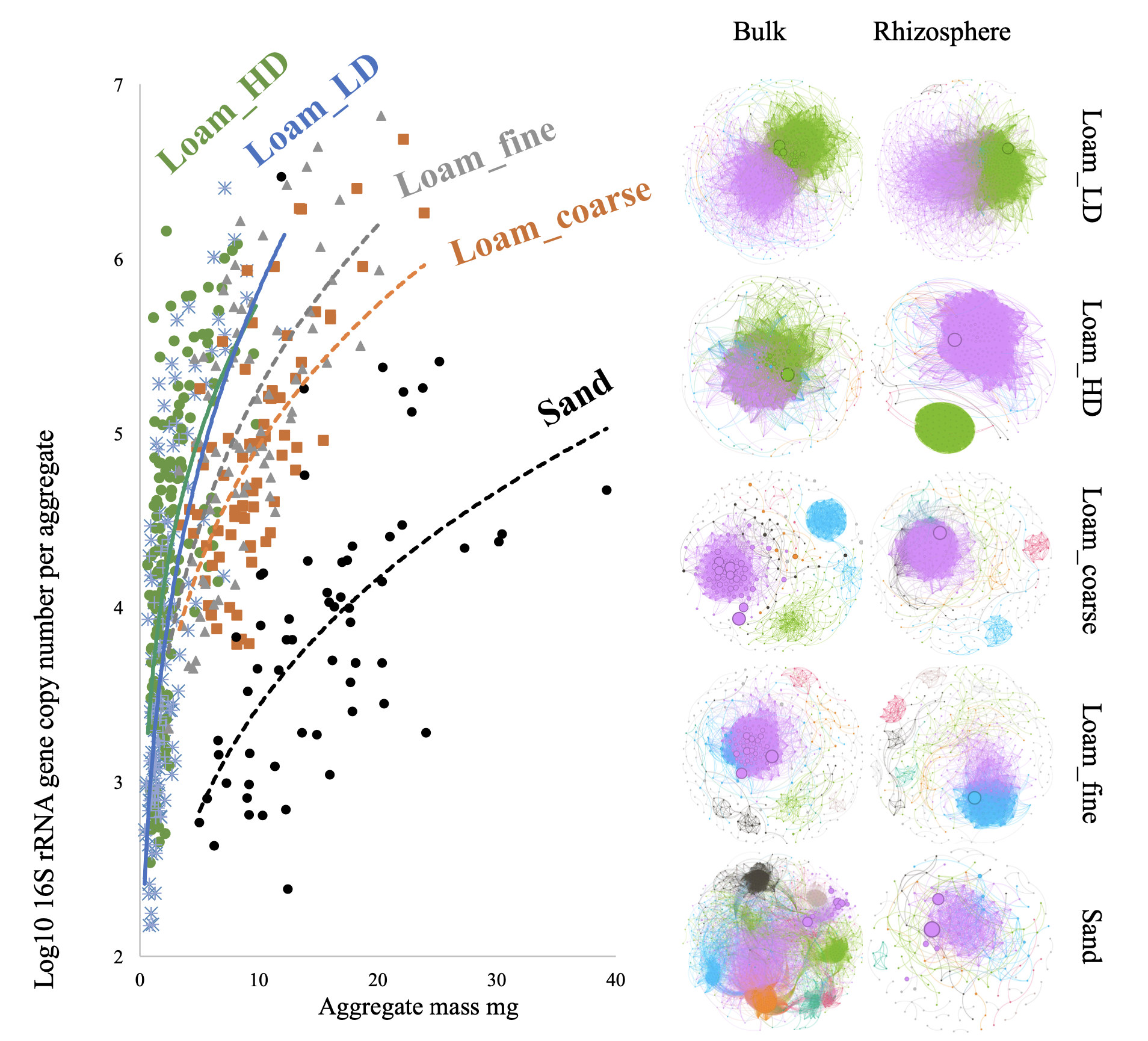
We investigated the effects of soil compaction and root-soil contact on the prokaryotic community structure within soil aggregates derived from the bulk soil and rhizosphere of the maize wild-type (WT) genotype. The study utilized soil column experiments to examine two soil bulk densities (low: 1.27 g cm⁻³ and high: 1.40 g cm⁻³) and three substrates (loam_coarse, loam_fine and sand). Analysis of 16S rRNA gene amplicons from DNA extracted from individual aggregates demonstrated that substrate type was the dominant factor influencing the prokaryotic community composition. The differentiation between bulk soil and rhizosphere communities was most pronounced in the loam_coarse substrate, suggesting that the bulk soil in this substrate experienced the least influence from maize roots compared to loam_fine and sandy substrates. Conversely, the differences between bulk soil and rhizosphere communities were least distinct in the sand substrate. Notably, the sand substrate exhibited the most densely connected prokaryotic co-occurrence network in the bulk soil and the least intensive network in the rhizosphere. The contrasting network patterns in sand suggest that microbial interactions in the bulk soil are primarily shaped by environmental constraints, promoting cooperation, whereas interactions in the rhizosphere are driven by resource availability, reducing the need for dense connectivity. These findings highlight how substrate properties and root exudation interact to structure microbial communities and influence their microbial networks. Soil density influenced prokaryotic taxa more prominently in the bulk soil relative to the rhizosphere. High soil density increased the abundance of prokaryotes within individual aggregates but did not significantly affect overall prokaryotic diversity. Moreover, high soil density amplified the differences in taxonomic abundance between bulk soil and rhizosphere communities. These findings suggest that soil density primarily influences microbial communities through physical and structural changes in the soil matrix. In the bulk soil, increased compaction creates spatially constrained environments that promote prokaryotic aggregation and abundance, while limiting diversity and connectivity to the rhizosphere. In contrast, the rhizosphere community remains more buffered against soil density effects due to root-derived inputs, resulting in amplified differences between the two compartments under high-density conditions. This highlights the dual role of soil density as both a physical barrier and an environmental filter, shaping microbial communities differently in bulk soil and the rhizosphere. Additionally, we investigated the prokaryotic community composition along a root-soil gradient (0–2 mm, 2–4 mm, 4–6 mm, and 6–8 mm from the root) associated with the rhizosphere of two maize genotypes: WT and root hair-defective mutant (rth3), using loam substrate soil column experiments. Sequence analysis of 16S rRNA gene amplicons from both DNA and RNA revealed that maize roots had no significant effect on the abundance or diversity of the prokaryotic community along the root-soil gradient. The lack of observable differentiation in the prokaryotic community along the root-soil gradient may indicate that either the roots exert a weak influence due to limited exudation or the loam substrate buffers microbial responses. Functional or strain-level shifts may still exist but require detection methods with higher taxonomic resolution. Further studies with more sensitive analyses and alternative substrates could clarify the underlying mechanisms.
Link to English scientific abstract
Link to German scientific abstract
