P8 - Root Genes
Spatial and temporal analysis of maize root gene expression to elucidate how the maize interacts with the rhizosphere microbiome and the environment
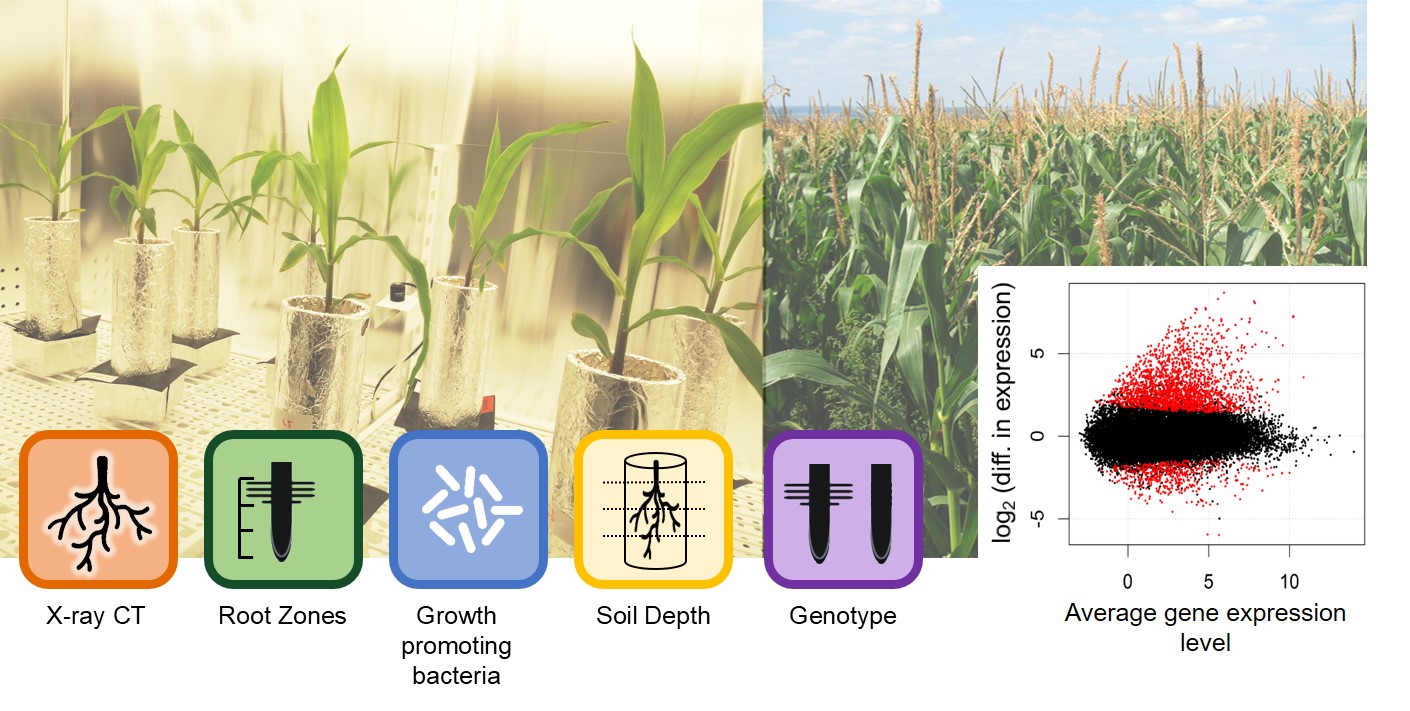
Plants shape rhizosphere communities by the deposition of root exudates, border cells and mucilage, competiting for nutrients and expressing immunity-related responses. In this project, we investigate plant-microbe interactions in the rhizosphere at the level of root gene expression in a spatial and temporal context. To obtain information how maize copes with soil compaction, drought stress and maize monoculture, wild type and root hair mutant maize are grown on loam and sand in soil column experiments under standardized conditions as well as in the field.
The main objective is to reveal mechanisms how maize roots adapt to changes in soil environment, and how the adaptations contribute to rhizosphere processes at a single root type and at root system levels. First, we investigate how soil compaction and increased soil contact of the root affects plant gene expression. Then, we analyze how water deficit, an important factor limiting maize production, is reflected in the abundances of plant immunity, nutrient transporter and root exudation related transcripts. We also want to dissect how the allocation level of carbon to root tips and rhizosphere is related to the response of maize to the rhizosphere microbiome during soil compaction. And finally, how the long-term development of soil structure and maize monoculture in the field is reflected by changes in the expression levels of genes for microbiome perception, defense signaling, exudation and nutrient uptake. The identification of the underlying gene expression patterns in synthesis with rhizosphere parameters investigated by collaborating research groups will advance a better understanding of the mechanisms through which plants shape and are shaped by the rhizosphere.
Outcome
1. A central experiment addressing the impact of a 7d drought on two maize genotypes (WT and rth3) revealed a strong influence of water limitation on root gene expression levels. Gene ontology enrichment analysis uncovered terms related to mineral element uptake, cell wall restructuring, defense, oxidative stress and secondary metabolism, representing a drought related gene expression pattern. An effect of water limitation and the genotype was observed on the community composition of rhizosphere ACC deaminase gene carrying (acdS+) bacteria, pointing out to an interaction between maize roots and the rhizosphere microbiome.
2. During a three-year field experiment using the same genotypes (WT, rth3) and two substrate (loam and sand) we investigated the impact of alternating annual precipitation (dry, moist, dry) and the role of precipitation history. In addition to a great effect of the factor year on gene expression levels we found differentially expressed genes related to drought and heat stress, mineral element uptake, exudation, immunity and defense, and cell wall restructuring in both dry years compared to the moist year. Nevertheless, we also found differences between both comparisons that can be attributed to the influence of precipitation in previous years and an associated adaptation to drought. We also observed an effect of the substrates that was more pronounced during dry years. Root stress enzyme activity showed different annual patterns between the substrates, likely influenced by an impact of repeated maize planting. Influenced by the same drivers, year and substrate had an effect on alpha- and beta-diversity of acdS+ bacteria in the rhizosphere, with an increase of gram-negative bacteria during the dry year. These results demonstrate the sensitivity of plant-microbe interactions to precipitation history and indicate an impact of repeated maize planting. The analysis will be continued in the future, by including one year before and one year after this 3-year period, focusing on the impact of repeated maize planting.
3. To investigate the impact of soil compaction and root-soil contact we joined two experiments, comparing two bulk densities and three soil textures on WT maize, respectively. For both experiments we only found a small number of differentially expressed genes. Changes in root anatomy due to soil compaction were low, but indicated an expansion of the cortical area. Investigation of rhizosphere acdS+ bacteria revealed a significant increase of Tetrasphaera under compacted soil, indicating an impact of soil compaction on plant-growth promoting bacteria. A detailed analysis of the results on root-soil contact follows.
Link to English scientific abstract
Link to German scientific abstract
