P16 - Rhizosphere Microbiome
Spatiotemporal organization of the rhizosphere microbiome shaped by external drivers
Soil ecosystem functions, as well as plant growth and health, are significantly influenced by the rhizosphere microbiome. The self-organization of the rhizosphere emerges from feedback loops involving roots, the microbiome, and soil.
In phase I, we investigated the microbiome associated with the rhizosphere of two maize genotypes (Root Hair), wild type (WT) and root hair defective mutant (rth3), in two different soil textures (Texture), loam and sand, and at different rooting depths (Depth). This was performed using both central soil column experiments in a growth chamber (SCE) and soil plot experiments in the field (SPE). During phase II, we further explored the spatiotemporal organization of the maize rhizosphere microbiome and external factors, particularly, seed inoculation with consortia of beneficial microorganisms (BMc) in drought conditions, how this affected plant development and the native rhizosphere microbiome.
The soil microbiome analysis was based on amplicon sequencing of the 16S rRNA gene fragments (bacteria/archaea) and ITS regions (fungi) amplified from total community DNAs isolated from soil samples, and based on culture dependent approaches.
Outcome
• X-ray computed tomography (XRCT) with a dose of 0.81 Gy was used to visualize maize root system architecture in the SCE (P8_SCE1). The XRCT had no effect on the bacterial diversity and composition in the WT maize rhizosphere grown in loam, observed one hour and 24 hours after exposure (
Ganther et al., 2020
).
• In another SCE (P8_SCE2), microbial colonization patterns along maize roots were analysed based on Texture, Depth, and Root Hair. The same key drivers influenced the rhizosphere bacteria, archaea and fungi (P16), microbial community carrying genes coded for 1-aminocyclopropane-1-carboxylate (P8), rhizosphere Protozoa community composition (P14), extracellular soil microbial enzyme activities (P13), and root gene expression (P8). Texture had the strongest impact, followed by Depth and Root Hair, indicating a close link between rhizosphere microbiome and plant gene expression (
Gebauer et al., 2021
;
Yim et al., 2022
).
• In SPE, similar patterns were observed, with Texture as the strongest divers. Changes in microbial community composition may be linked to exudation rates (amino acid, carboxylic acid, and sugar found by P11) that favour fast-growing microbes reducing diversity at the vegetative stage and diversity recovered at the reproductive stage when exudation rates are low again. Changes in substrate availability over time, microbe-microbe interactions and top-down control by microbial predators are likely contributed to a more stable and diverse rhizosphere microbial community at the reproductive stage (Oburger et al., 2025, MS in preparation). Additionally, we also evaluated the SPE, whether maize monoculture for five years (2019 - 2023) led to replant depression. Variations in bacterial/archaeal community composition over time may be driven by both monoculture and precipitation, as indicated by changes in the relative abundance of responders belonging to Gram-positive and -negative bacteria. Among the top most relative abundance bacterial ASVs, none of them were linked to maize replant depression. However, thorough assessments of bacterial/archaeal and fungal communities are ongoing.
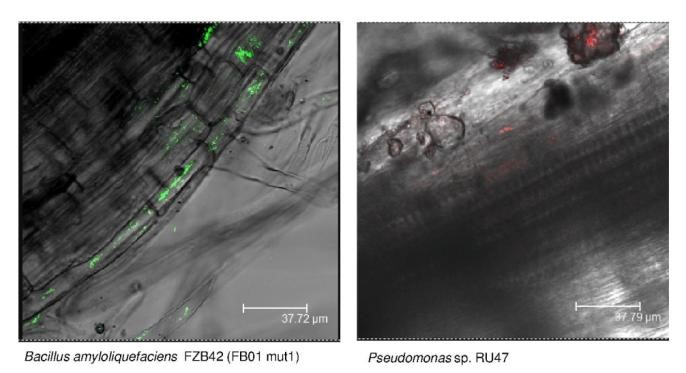
• In SCE_drought (P6, P11, P16), seven days of drought stress significantly shifted microbial beta-diversity in 22-days-old WT and rth3 rhizospheres, with fungi showing stronger effects than bacteria and archaea (Hartwig et al., 2025, in print
). Under greenhouse conditions (P16), six beneficial bacterial isolates obtained from maize (rhizosphere or rhizoplane) and selected due to their potential drought-mitigating traits and dominant abundance in the rhizosphere (
Yim et al., 2022
;
He et al., 2024
) were tested for drought-mitigation. WT seed inoculation with BMc improved plant growth under both well-watered and drought conditions in loam without disrupting native microbial communities (
Yim et al., 2025
). These fundings highlight the potential of seed inoculation with BMc to enhance drought tolerance in maize.
Link to English scientific abstract
Link to German scientific abstract
