P15 - Mycorrhizosphere
Effect of genetic perturbations on the spatial heterogeneity and co-occurrence patterns in the microbiota of the maize mycorrhizosphere in interaction with two different soils
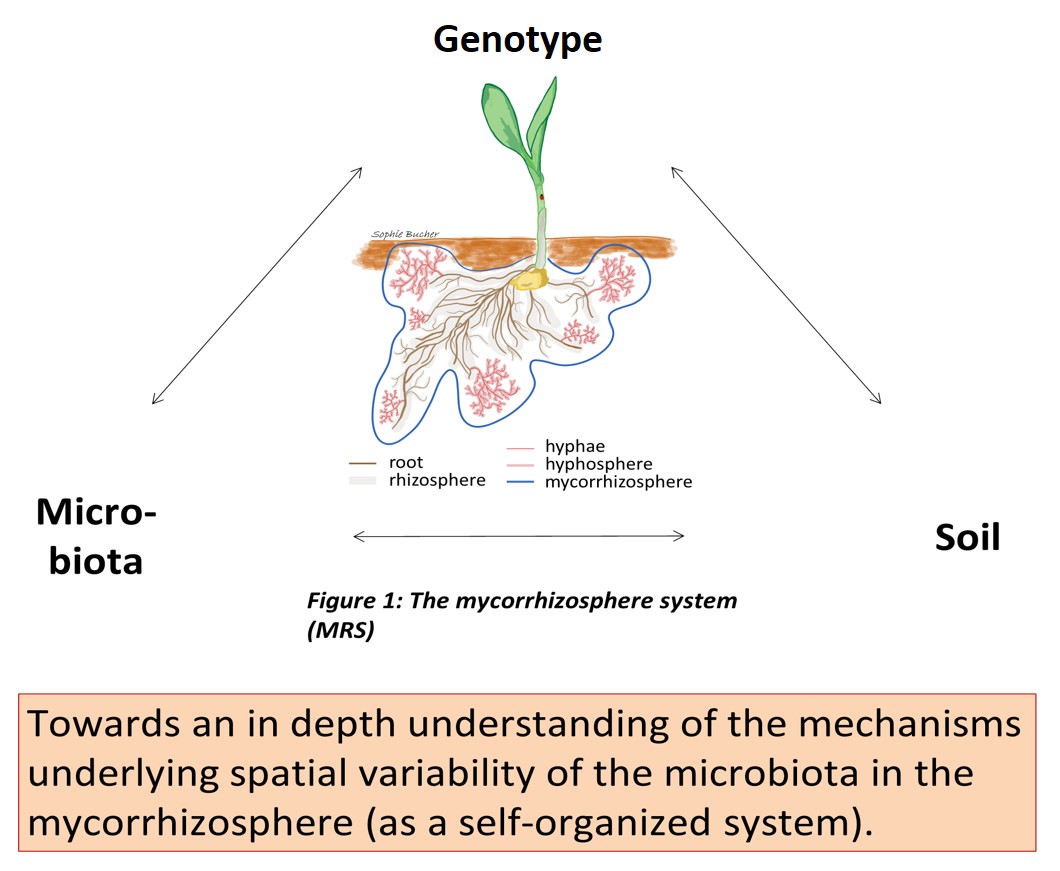
A huge variety of microbes play important roles in thriving of maize (Zea mays) in agricultural soils. Arbuscular mycorrhizal fungi (AMF) belong to the maize root microbiota and deliver a large proportion of mineral nutrients (e.g. phosphate) to the plant through their hyphal network. At the present, nutritional and metabolic interactions between plants, AMF and their microbiota, and how they affect soil properties, the mycorrhizospheric microbiota and plant growth, is far from being fully understood. To elucidate how plant factors shape the interdependency between root, microbiota and soil, we investigated the microbiome variation in the mycorrhizosphere, comprising the root endosphere, rhizosphere, hyphoplane (surface of fungal hyphae) and hyphosphere, of two maize inbred lines (B73, OH43) and two maize mutants affected in root hair formation and mycorrhizal phosphate uptake (rth3, pht1;6) grown under varying phosphorus (P) and nitrogen (N) conditions in three consecutive field studies using automated ribosomal intergenic spacer analysis (ARISA) and amplicon sequencing.
To assess the functional evolution and adaptability of the mycorrhizosphere microbiome, we used host-mediated microbiome engineering (HMME), which exploits host phenotype to indirectly select microbiomes through cyclic propagation. Wildtype maize (B73) and mutant lines (pht1;6) impaired in the Mycorrhizal Phosphate Uptake (MPU) pathway undergo ten selection cycles under phosphorus-deficient conditions. By assessing plant biomass, leaf nutrient content, and photosynthetic activity, along with rhizosphere 16S profiling, the study aims to identify shifts in microbial community composition driven by specific plant traits. We are establishing a rich collection of fully indexed isolates of mycorrhiza-associated microbes to establish synthetic communities (SynComs) of reduced complexity representing the genetic diversity of the natural microbial community of mycorrhizal maize. Germ-free maize (wild type and mutants) will be re-populated with these microbiota from the field and greenhouse study, to assess their impact on host fitness and performance and cross-kingdom species interactions. Furthermore, SynComs including fungal and bacterial isolates with keystone species from mycorrhizal maize roots will enable studying the biological role(s) of individual mycorrhizospheric microorganisms and underlying molecular mechanisms under controlled and reproducible conditions.
Outcome phase 1 – Microbiome variation in the mycorrhizosphere
We developed a half-automated sieving and sucrose centrifugation (SSC) method to efficiently obtain the extraradical fungal hyphae (ERH) with attached microbes (hyphoplane) from natural loamy field soil. Amplicon sequencing revealed that the bacterial and fungal communities inhabiting the root endosphere and rhizosphere differed remarkably in diversity and structure from those found in the extraradical hyphoplane and hyphosphere. Soil P and N fertilization, and to a lesser extent plant genotype, affected the microbial communities throughout all mycorrhizal compartments, indicating that changes in the plant genotype are transmitted to community assembly by ERH. The knock-out of the mycorrhizal phosphate transporter PHT1;6 had the strongest effects on the growth and nutrient content of maize and on the composition of the bacterial community on the hyphoplane among the four plant genotypes investigated. The loss of MPU more strongly shaped fungal and bacterial communities in the root endosphere than in the rhizosphere, hyphoplane, and hyphosphere, but left ERH growth unchanged.
Outcome phase 2 - Functional evolution and adaptability of the mycorrhizosphere microbiome
In the first selection cycle of the HMME experiment, the shoot fresh weight of the best and worst growing plants is similar. However, in cycle ten, the best-growing plants have a significantly higher shoot fresh weight than the worst-growing plants in both genotypes. While artificial selection successfully increased shoot fresh weight in the best-growing plants, it did not lead to a decrease in the worst-growing plants. Our results show that the rhizosphere bacterial community of young maize roots is strongly affected by artificial selection rather than microbial aging or environmental factors. Additionally, B73 and pht1;6 genotypes shape the rhizosphere bacterial community in early growth stage, and this influence is stronger in late selection cycles. This suggests that artificial evolution is selecting for a specialized and potentially functional microbial community early in the plant´s life cycle. Furthermore, both positive and negative artificial selection on shoot biomass is associated with changes in the rhizosphere bacterial community. The increased abundance of Azospirillum, Burkholderia and Sphingomonadaceae in later selection cycles highlights their importance in nutrient acquisition and plant performance. Further studies will assess their interactions to uncover cooperative or synergistic relationships that drive microbiome.
Link to English scientific abstract
Link to German scientific abstract
