Project participated only in SPP-Phase 1
P20 - Biochemical Gradients
Bridging biochemical gradients to elucidate key processes governing root -microbiome-soil interactions
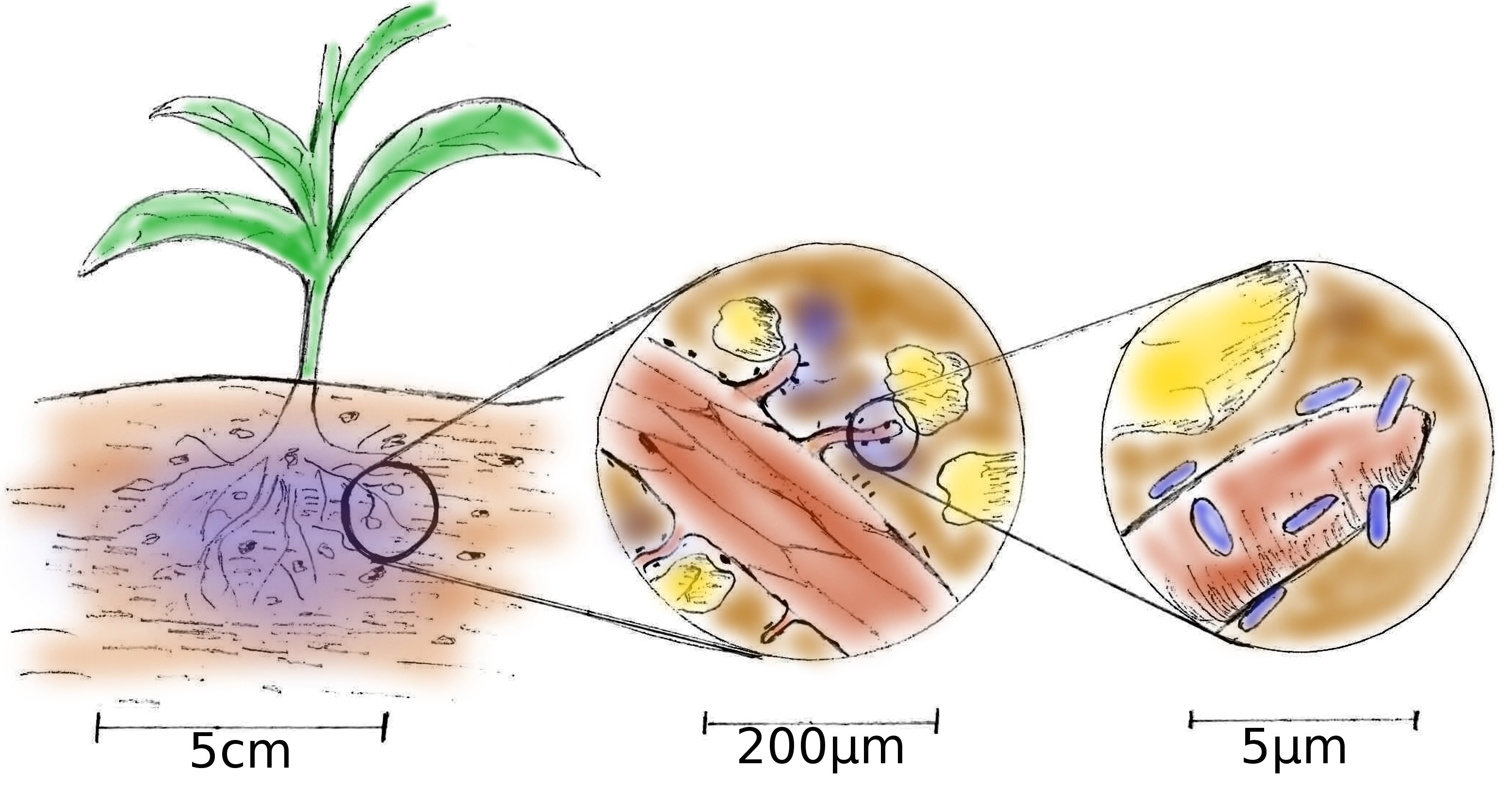
The rhizosphere is the boundary between plant roots and soil and is a highly complex and dynamic system whose self-organization, function and ecology is governed by spatio-temporal chemical gradients across scales ranging from nm to cm. These gradients result from an active and continuous interplay between roots, soil components and the microbiome. The plant roots act as the key source of organic carbon which they provide as root exudates. These exudates are the main energy source that fuels the rhizosphere-microbiome. In turn, the microbes catalyse various chemical reactions that make nutrients, e.g. nitrogen, phosphorous and metals, available to the plant and mineralize organic matter. Thus, heterogenic patterns of microbial colonies, mineral particles and the distribution of carbon and nutrients govern the self-organisation of the rhizosphere.
Several methods such as X-ray CT, Neutron imaging and MRI/NMR, to name some, have been developed to image the structure and to measure the composition of the rhizosphere. However, their spatial resolution is not sufficient to measure chemical gradients on the scale of the microbial cells attached to soil aggregates, mycorrhiza, roots and root hairs. On the other hand, the self-organisation of the rhizosphere can only be understood by elucidating the spatial dimensions of individual microbial processes. To fill this gap this project aims to identify, quantify and visualise biotic and abiotic processes across scales and at the level of a single cell, sub-cellular component or single soil aggregate. The biogeochemical gradients of C, N, P and micro-nutrients (e.g. B) at different scales ranging from a cm level representative for the root in a soil matrix to a sub μm level representing the habitat of a single cell in a biofilm interacting with soil matrix or the surface of a root / root hair are imaged by means of correlative chemical microscopy. For that, rhizosphere samples are embedded in resin and cleaved or sectioned to obtain flat surfaces. Fluorescense in-situ hybridisation is employed to determine the identity of microbial cells in the rhizosphere-microbiome. Stable-isotope labelling (2H, 13C, 15N) in conjunction with secondary ion-mass spectroscopy (nanoSIMS and time-of-flight SIMS) as well as laser-ablation inductively-coupled plasma mass-spectroscopy is used to trace biological activity and fluxes of carbon and nutrients. Scanning electron microscopy with energy-dispersive X-ray spectroscopy is employed for elemental analysis at high-resolution. Scanning helium-ion microscopy is used for high-resolution imaging.
Link to English scientific abstract
Link to German scientific abstract
