Research
The Cell4Chem project aims to enhance the power of microbial communities in a sustainable, circular biobased economy. For this purpose, Cell4Chem will engineer bacterial communities to enable the use of lignocellulosic feedstock for the efficient production of specialty chemicals. This will expand the range of substrate for the carboxylate platform. To improve the applicability of microbial communities in future biotechnological processes, Cell4Chem takes a multidisciplinary approach.
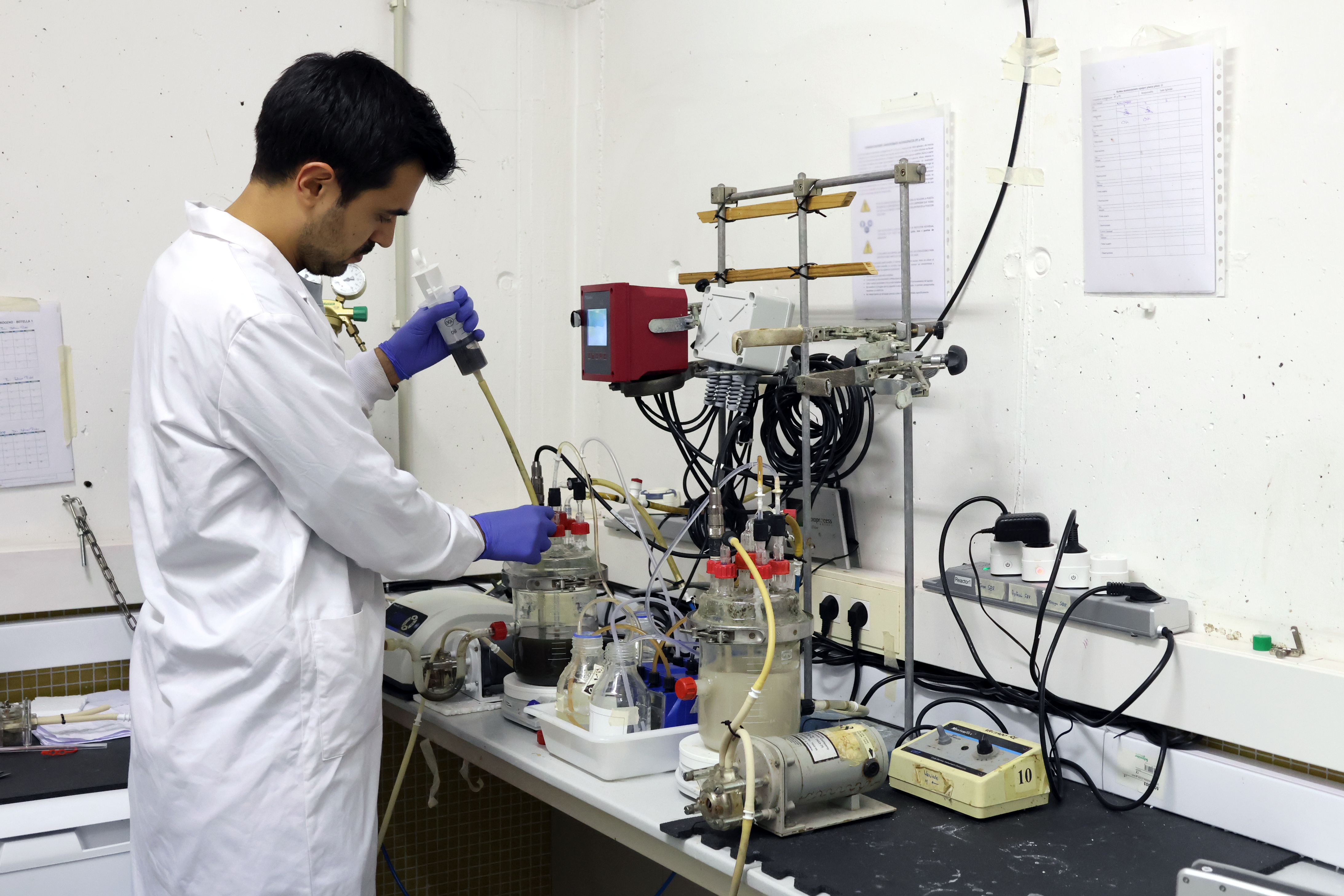
Cell4Chem comprises the development and assessment of an anaerobic fermentation process for the production of green chemicals, mainly caproate, from lignocellulosic biomass, thereby expanding the applicability of the carboxylate platform. Two bottlenecks in the anaerobic fermentation of lignocellulose are in the focus, i.e. hydrolysis of cellulose and internal lactate production, to fuel microbial chain elongation of short-chain carboxylates and enhance productivity and yield. The metabolic potential of genetically modified bacteria will be assessed in constructed consortia together with wildtype strains and enrichment cultures including chain elongation bacteria.
A Synthetic Biology approach will be used for genetic engineering of lactic acid bacteria to provide it with the ability to degrade and use cellulose and increase the production of lactate. Further bacteria will be engineered to support the lactate production. Genetic parts will be prepared in line with synthetic biology standards, which will greatly facilitate cloning and selection of optimal plasmid constructs, as well as exchange between laboratories. Species-specific codon optimisation and the use of synthetic genes will be considered. All genetic designs will be specified in Synthetic Biology Open Language (SBOL), visualised using SBOL Designer software and shared in the SynBioHub repository.

Bioinformatic tools will be used to analyze meta-omics data and reveal the structure and activity of microbial consortia. Consortia structure will be monitored with amplicon sequencing data of 16S rRNA genes. For promising consortia, metagenomics analysis will be used to reconstruct bacterial genomes and assess their metabolic functions. To that, both short- and long-read sequencing strategies will be applied aiming at high quality and contiguous genomes. Metaproteomics will be used to investigate microbial consortia’s activity and key enzymes of the process. Together, meta-omics data will provide a deeper understanding of the carboxylate production from lignocellulosic biomass.
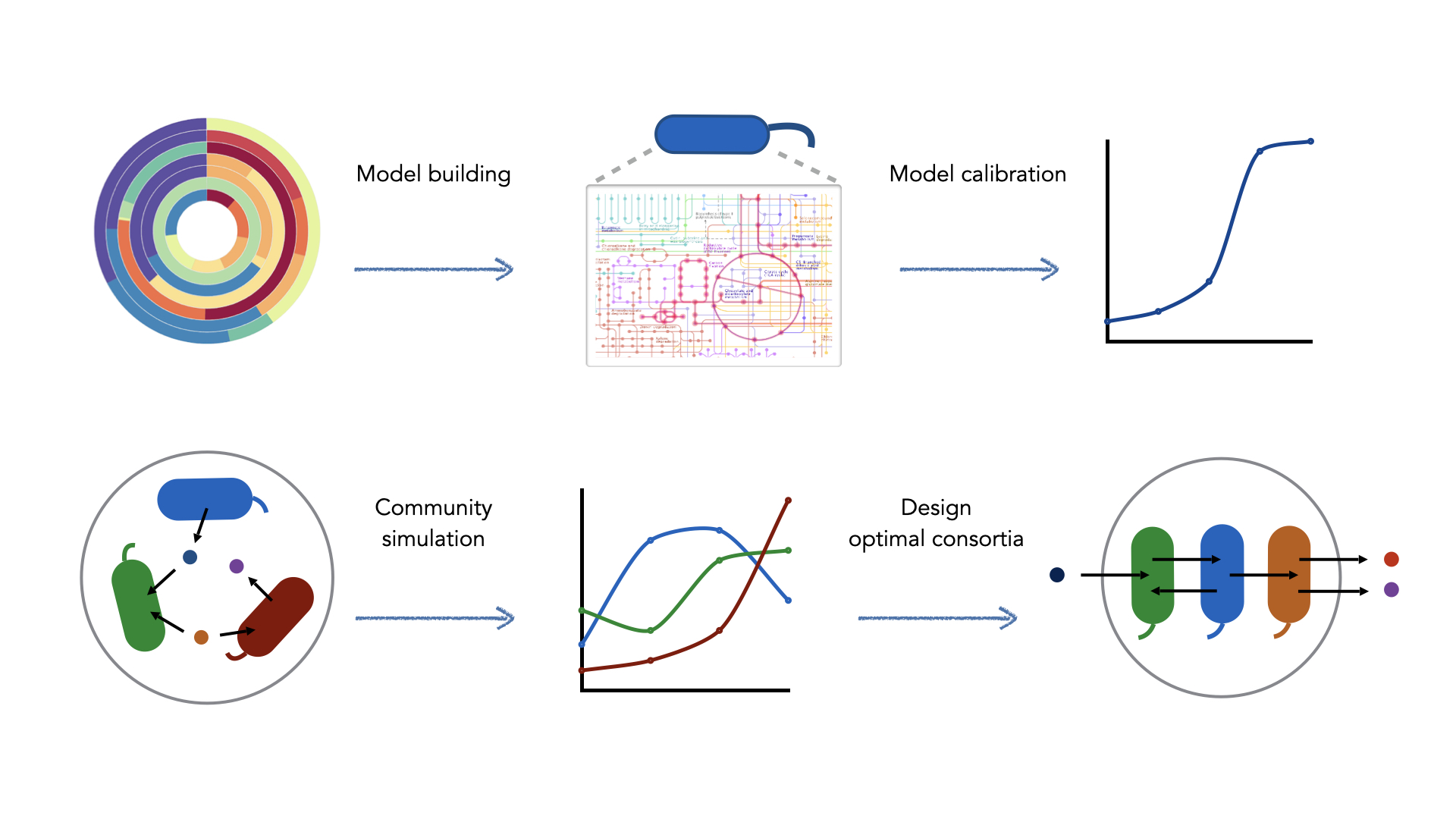
We use a Systems Biology approach using state-of-the-art methods to model microbial consortia combining genome-scale metabolic modelling and meta-omics data integration. We use CarveMe to build genome-scale metabolic models (GEMs) for all bacterial strains used in the project from reference genomes and MAGs. These strains are combined into multiple consortia and their metabolic phenotypes are simulated using μbialSim. Integration of omics data are used to elucidate metabolic cross-feeding interactions and identify potential bottlenecks. DesignMC will be used to identify the most promising community combinations and to guide the design of optimal microbial communities that reach the highest product yields.